A Fast and Accessible Method for the Isolation of RNA, DNA, and Protein To Facilitate the Detection of SARS-CoV-2
- PMID: 33293367
- PMCID: PMC8092744
- DOI: 10.1128/JCM.02403-20
A Fast and Accessible Method for the Isolation of RNA, DNA, and Protein To Facilitate the Detection of SARS-CoV-2
Abstract
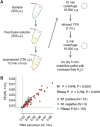
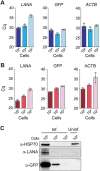
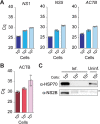
Management of the coronavirus disease 2019 (COVID-19) pandemic requires widespread testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A main limitation for widespread SARS-CoV-2 testing is the global shortage of essential supplies, among them RNA extraction kits. The need for commercial RNA extraction kits places a bottleneck on tests that detect SARS-CoV-2 genetic material, including PCR-based reference tests. Here, we propose an alternative method we call PEARL (
Keywords: DNA and protein; SARS-CoV-2; field deployable; rapid isolation of RNA; virus detection.
Copyright © 2021 Ponce-Rojas et al.
Figures
References
-
- CDC. 2019. Novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. CDC, Atlanta, GA. https://www.fda.gov/media/134922/download. - PMC - PubMed
-
- Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Muenker MC, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Cruz CSD, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND. 2020. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol 5:1299–1305. doi: 10.1038/s41564-020-0761-6. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

