Inductive inference of gene regulatory network using supervised and semi-supervised graph neural networks
- PMID: 33294129
- PMCID: PMC7677691
- DOI: 10.1016/j.csbj.2020.10.022
Inductive inference of gene regulatory network using supervised and semi-supervised graph neural networks
Abstract
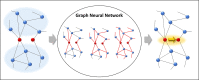
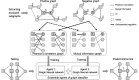
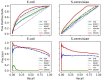
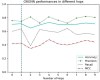
Discovering gene regulatory relationships and reconstructing gene regulatory networks (GRN) based on gene expression data is a classical, long-standing computational challenge in bioinformatics. Computationally inferring a possible regulatory relationship between two genes can be formulated as a link prediction problem between two nodes in a graph. Graph neural network (GNN) provides an opportunity to construct GRN by integrating topological neighbor propagation through the whole gene network. We propose an end-to-end gene regulatory graph neural network (GRGNN) approach to reconstruct GRNs from scratch utilizing the gene expression data, in both a supervised and a semi-supervised framework. To get better inductive generalization capability, GRN inference is formulated as a graph classification problem, to distinguish whether a subgraph centered at two nodes contains the link between the two nodes. A linked pair between a transcription factor (TF) and a target gene, and their neighbors are labeled as a positive subgraph, while an unlinked TF and target gene pair and their neighbors are labeled as a negative subgraph. A GNN model is constructed with node features from both explicit gene expression and graph embedding. We demonstrate a noisy starting graph structure built from partial information, such as Pearson's correlation coefficient and mutual information can help guide the GRN inference through an appropriate ensemble technique. Furthermore, a semi-supervised scheme is implemented to increase the quality of the classifier. When compared with established methods, GRGNN achieved state-of-the-art performance on the DREAM5 GRN inference benchmarks. GRGNN is publicly available at https://github.com/juexinwang/GRGNN.
Keywords: Gene regulatory; Graph neural networks; Inductive learning; Machine learning.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
Predicting gene regulatory links from single-cell RNA-seq data using graph neural networks.Brief Bioinform. 2023 Sep 22;24(6):bbad414. doi: 10.1093/bib/bbad414. Brief Bioinform. 2023. PMID: 37985457 Free PMC article.
-
Graph attention network for link prediction of gene regulations from single-cell RNA-sequencing data.Bioinformatics. 2022 Sep 30;38(19):4522-4529. doi: 10.1093/bioinformatics/btac559. Bioinformatics. 2022. PMID: 35961023
-
An inductive graph neural network model for compound-protein interaction prediction based on a homogeneous graph.Brief Bioinform. 2022 May 13;23(3):bbac073. doi: 10.1093/bib/bbac073. Brief Bioinform. 2022. PMID: 35275993 Free PMC article. Review.
-
Attribute-driven streaming edge partitioning with reconciliations for distributed graph neural network training.Neural Netw. 2023 Aug;165:987-998. doi: 10.1016/j.neunet.2023.06.026. Epub 2023 Jun 28. Neural Netw. 2023. PMID: 37467586 Review.
Cited by
-
MIGGRI: A multi-instance graph neural network model for inferring gene regulatory networks for Drosophila from spatial expression images.PLoS Comput Biol. 2023 Nov 8;19(11):e1011623. doi: 10.1371/journal.pcbi.1011623. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 37939200 Free PMC article.
-
Inferring Gene Regulatory Networks From Single-Cell Transcriptomic Data Using Bidirectional RNN.Front Oncol. 2022 May 26;12:899825. doi: 10.3389/fonc.2022.899825. eCollection 2022. Front Oncol. 2022. PMID: 35692809 Free PMC article.
-
DeepFGRN: inference of gene regulatory network with regulation type based on directed graph embedding.Brief Bioinform. 2024 Mar 27;25(3):bbae143. doi: 10.1093/bib/bbae143. Brief Bioinform. 2024. PMID: 38581416 Free PMC article.
-
Inference of Gene Regulatory Networks Based on Multi-view Hierarchical Hypergraphs.Interdiscip Sci. 2024 Jun;16(2):318-332. doi: 10.1007/s12539-024-00604-3. Epub 2024 Feb 11. Interdiscip Sci. 2024. PMID: 38342857
-
Computational approaches to understand transcription regulation in development.Biochem Soc Trans. 2023 Feb 27;51(1):1-12. doi: 10.1042/BST20210145. Biochem Soc Trans. 2023. PMID: 36695505 Free PMC article. Review.
References
-
- Aliferis C.F., Statnikov A., Tsamardinos I., Mani S., Koutsoukos X.D. Local causal and markov blanket induction for causal discovery and feature selection for classification Part i: algorithms and empirical evaluation. J Mach Learn Res. 2010;11:171–234.
-
- Bassett G.W., Tam M.-Y.-S., Knight K. Quantile models and estimators for data analysis. In: Dutter R., Filzmoser P., Gather U., Rousseeuw P.J., editors. Developments in Robust Statistics. Physica-Verlag HD; Heidelberg: 2003. pp. 77–87. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
