Integration of mechanistic immunological knowledge into a machine learning pipeline improves predictions
- PMID: 33294774
- PMCID: PMC7720904
- DOI: 10.1038/s42256-020-00232-8
Integration of mechanistic immunological knowledge into a machine learning pipeline improves predictions
Abstract
The dense network of interconnected cellular signalling responses that are quantifiable in peripheral immune cells provides a wealth of actionable immunological insights. Although high-throughput single-cell profiling techniques, including polychromatic flow and mass cytometry, have matured to a point that enables detailed immune profiling of patients in numerous clinical settings, the limited cohort size and high dimensionality of data increase the possibility of false-positive discoveries and model overfitting. We introduce a generalizable machine learning platform, the immunological Elastic-Net (iEN), which incorporates immunological knowledge directly into the predictive models. Importantly, the algorithm maintains the exploratory nature of the high-dimensional dataset, allowing for the inclusion of immune features with strong predictive capabilities even if not consistent with prior knowledge. In three independent studies our method demonstrates improved predictions for clinically relevant outcomes from mass cytometry data generated from whole blood, as well as a large simulated dataset. The iEN is available under an open-source licence.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


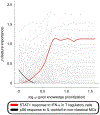
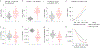

References
-
- Rieckmann JC et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol 18, 583–593 (2017). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
