Designed Metal-ATCUN Derivatives: Redox- and Non-redox-Based Applications Relevant for Chemistry, Biology, and Medicine
- PMID: 33294799
- PMCID: PMC7701195
- DOI: 10.1016/j.isci.2020.101792
Designed Metal-ATCUN Derivatives: Redox- and Non-redox-Based Applications Relevant for Chemistry, Biology, and Medicine
Abstract
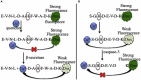
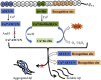
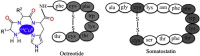
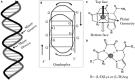
The designed "ATCUN" motif (amino-terminal copper and nickel binding site) is a replica of naturally occurring ATCUN site found in many proteins/peptides, and an attractive platform for multiple applications, which include nucleases, proteases, spectroscopic probes, imaging, and small molecule activation. ATCUN motifs are engineered at periphery by conjugation to recombinant proteins, peptides, fluorophores, or recognition domains through chemically or genetically, fulfilling the needs of various biological relevance and a wide range of practical usages. This chemistry has witnessed significant growth over the last few decades and several interesting ATCUN derivatives have been described. The redox role of the ATCUN moieties is also an important aspect to be considered. The redox potential of designed M-ATCUN derivatives is modulated by judicious choice of amino acid (including stereochemistry, charge, and position) that ultimately leads to the catalytic efficiency. In this context, a wide range of M-ATCUN derivatives have been designed purposefully for various redox- and non-redox-based applications, including spectroscopic probes, target-based catalytic metallodrugs, inhibition of amyloid-β toxicity, and telomere shortening, enzyme inactivation, biomolecules stitching or modification, next-generation antibiotic, and small molecule activation.
Keywords: Biochemistry; Chemistry; Inorganic Chemistry; Medical Biochemistry.
Figures










Similar articles
-
Improved bioactivity of antimicrobial peptides by addition of amino-terminal copper and nickel (ATCUN) binding motifs.ChemMedChem. 2014 Aug;9(8):1892-901. doi: 10.1002/cmdc.201402033. Epub 2014 May 6. ChemMedChem. 2014. PMID: 24803240 Free PMC article.
-
Metal-binding and redox properties of substituted linear and cyclic ATCUN motifs.J Inorg Biochem. 2014 Oct;139:65-76. doi: 10.1016/j.jinorgbio.2014.06.004. Epub 2014 Jun 12. J Inorg Biochem. 2014. PMID: 24980953 Free PMC article.
-
Hybrid peptide ATCUN-sh-Buforin: Influence of the ATCUN charge and stereochemistry on antimicrobial activity.Biochimie. 2015 Jun;113:143-55. doi: 10.1016/j.biochi.2015.04.008. Epub 2015 Apr 17. Biochimie. 2015. PMID: 25891844
-
Kinetics of Cu(II) complexation by ATCUN/NTS and related peptides: a gold mine of novel ideas for copper biology.Dalton Trans. 2021 Dec 20;51(1):14-26. doi: 10.1039/d1dt02878b. Dalton Trans. 2021. PMID: 34816848 Review.
-
Target-directed catalytic metallodrugs.Braz J Med Biol Res. 2013 Jun;46(6):465-85. doi: 10.1590/1414-431X20133086. Epub 2013 Jul 2. Braz J Med Biol Res. 2013. PMID: 23828584 Free PMC article. Review.
Cited by
-
Restoring cellular copper homeostasis in Alzheimer disease: a novel peptide shuttle is internalized by an ATP-dependent endocytosis pathway involving Rab5- and Rab14-endosomes.Front Mol Biosci. 2024 Apr 5;11:1355963. doi: 10.3389/fmolb.2024.1355963. eCollection 2024. Front Mol Biosci. 2024. PMID: 38645276 Free PMC article.
-
Boosting AMPs' Power: From Structural Engineering to Nanotechnology-Based Delivery.Molecules. 2025 Jul 15;30(14):2979. doi: 10.3390/molecules30142979. Molecules. 2025. PMID: 40733245 Free PMC article. Review.
-
A Copper-Binding Peptide with Therapeutic Potential against Alzheimer's Disease: From the Blood-Brain Barrier to Metal Competition.ACS Chem Neurosci. 2025 Jan 15;16(2):241-261. doi: 10.1021/acschemneuro.4c00796. Epub 2024 Dec 26. ACS Chem Neurosci. 2025. PMID: 39723808 Free PMC article.
-
Why the Ala-His-His Peptide Is an Appropriate Scaffold to Remove and Redox Silence Copper Ions from the Alzheimer's-Related Aβ Peptide.Biomolecules. 2022 Sep 20;12(10):1327. doi: 10.3390/biom12101327. Biomolecules. 2022. PMID: 36291536 Free PMC article.
-
A Portable, Encapsulated Microbial Whole-Cell Biosensing System for the Detection of Bioavailable Copper (II) in Soil.Microchem J. 2023 Oct;193:109088. doi: 10.1016/j.microc.2023.109088. Epub 2023 Jul 17. Microchem J. 2023. PMID: 37982106 Free PMC article.
References
-
- Aboul-ela F., Varani G. Recognition of HIV-1 TAR RNA by tat protein and tat-derived peptides. J. Mol. Struct. 1998;423:29–39.
-
- Agbale C.M., Cardoso M.H., Galyuon I.K., Franco O.L. Designing metallodrugs with nuclease and protease activity. Metallomics. 2016;8:1159–1169. - PubMed
-
- Alexander J.L., Yu Z., Cowan J.A. Amino terminal copper and nickel binding motif derivatives of ovispirin-3 display increased antimicrobial activity via lipid oxidation. J. Med. Chem. 2017;60:10047–10055. - PubMed
-
- Alexander J.L., Thompson Z., Yu Z., Cowan J.A. Antimicrobial metallopeptides. ACS Chem. Biol. 2018;13:844–853. - PubMed
-
- Alexander J.L., Thompson Z., Yu Z., Cowan J.A. Cu-ATCUN derivatives of Sub5 exhibit enhanced antimicrobial activity via multiple modes of action. ACS. Chem. Biol. 2019;14:449–458. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

