Divergent Nodes of Non-autonomous UPRER Signaling through Serotonergic and Dopaminergic Neurons
- PMID: 33296657
- PMCID: PMC8820220
- DOI: 10.1016/j.celrep.2020.108489
Divergent Nodes of Non-autonomous UPRER Signaling through Serotonergic and Dopaminergic Neurons
Abstract
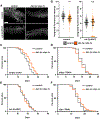
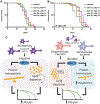
In multicellular organisms, neurons integrate a diverse array of external cues to affect downstream changes in organismal health. Specifically, activation of the endoplasmic reticulum (ER) unfolded protein response (UPRER) in neurons increases lifespan by preventing age-onset loss of ER proteostasis and driving lipid depletion in a cell non-autonomous manner. The mechanism of this communication is dependent on the release of small clear vesicles from neurons. We find dopaminergic neurons are necessary and sufficient for activation of cell non-autonomous UPRER to drive lipid depletion in peripheral tissues, whereas serotonergic neurons are sufficient to drive protein homeostasis in peripheral tissues. These signaling modalities are unique and independent and together coordinate the beneficial effects of neuronal cell non-autonomous ER stress signaling upon health and longevity.
Keywords: UPRER; aging; non-autonomous signaling; stress response.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Chase DL, Pepper JS, and Koelle MR (2004). Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat. Neurosci 7, 1096–1103. - PubMed

