SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation
- PMID: 33296685
- PMCID: PMC7680029
- DOI: 10.1016/j.immuni.2020.11.009
SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation
Abstract
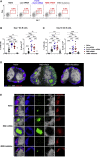
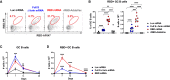
The deployment of effective vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical to eradicate the coronavirus disease 2019 (COVID-19) pandemic. Many licensed vaccines confer protection by inducing long-lived plasma cells (LLPCs) and memory B cells (MBCs), cell types canonically generated during germinal center (GC) reactions. Here, we directly compared two vaccine platforms-mRNA vaccines and a recombinant protein formulated with an MF59-like adjuvant-looking for their abilities to quantitatively and qualitatively shape SARS-CoV-2-specific primary GC responses over time. We demonstrated that a single immunization with SARS-CoV-2 mRNA, but not with the recombinant protein vaccine, elicited potent SARS-CoV-2-specific GC B and T follicular helper (Tfh) cell responses as well as LLPCs and MBCs. Importantly, GC responses strongly correlated with neutralizing antibody production. mRNA vaccines more efficiently induced key regulators of the Tfh cell program and influenced the functional properties of Tfh cells. Overall, this study identifies SARS-CoV-2 mRNA vaccines as strong candidates for promoting robust GC-derived immune responses.
Keywords: COVID-19; SARS-CoV-2; T follicular helper cells; germinal center B cells; germinal centers; mRNA vaccines; neutralizing antibodies.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that Drew Weissman is named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins. Drew Weissman and Norbert Pardi are also named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles (LNPs) as a vaccine platform. We have disclosed those interests fully to the University of Pennsylvania, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents. Ying K. Tam and Steven H.Y. Fan are employees of Acuitas Therapeutics, a company involved in the development of mRNA-LNP therapeutics. Ying Tam is named on patents that describe LNPs for delivery of nucleic acid therapeutics including mRNA and the use of modified mRNA in LNPs as a vaccine platform.
Figures






References
-
- Baumjohann D., Okada T., Ansel K.M. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 2011;187:2089–2092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

