Low-Avidity CD4+ T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19
- PMID: 33296686
- PMCID: PMC7689350
- DOI: 10.1016/j.immuni.2020.11.016
Low-Avidity CD4+ T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19
Abstract
CD4+ T cells reactive against SARS-CoV-2 can be found in unexposed individuals, and these are suggested to arise in response to common cold coronavirus (CCCoV) infection. Here, we utilized SARS-CoV-2-reactive CD4+ T cell enrichment to examine the antigen avidity and clonality of these cells, as well as the relative contribution of CCCoV cross-reactivity. SARS-CoV-2-reactive CD4+ memory T cells were present in virtually all unexposed individuals examined, displaying low functional avidity and multiple, highly variable cross-reactivities that were not restricted to CCCoVs. SARS-CoV-2-reactive CD4+ T cells from COVID-19 patients lacked cross-reactivity to CCCoVs, irrespective of strong memory T cell responses against CCCoV in all donors analyzed. In severe but not mild COVID-19, SARS-CoV-2-specific T cells displayed low functional avidity and clonality, despite increased frequencies. Our findings identify low-avidity CD4+ T cell responses as a hallmark of severe COVID-19 and argue against a protective role for CCCoV-reactive T cells in SARS-CoV-2 infection.
Keywords: ARTE; COVID-19; SARS-CoV-2; T cell cross-reactivity; antigen-reactive T cell enrichment; antigen-specific T cells; common cold coronavirus; human coronavirus; pre-existing memory.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests P.B. and A.S. are consultants of Miltenyi Biotec, who own IP rights concerning parts of the ARTE technology. M.J.G.T.V. has received research grants from 3M, Astellas Pharma, Biontech, DaVolterra, Evonik, Gilead Sciences, Glycom, Immunic, MaaT Pharma, Merck/MSD, Organobalance, Seres Therapeutics, and Takeda Pharmaceutical; speakers fees from Astellas Pharma, Basilea, Gilead Sciences, Merck/MSD, Organobalance, and Pfizer; and consultation fees from Alb Fils Kliniken GmbH, Arderypharm, Astellas Pharma, DaVolterra, Farmak International Holding GmbH, Ferring, Immunic AG, MaaT Pharma, Merck/MSD, and SocraTec R&D GmbH. F.L. discloses speaker honoraria from Grifols, Teva, Biogen, Bayer, Roche, Novartis, and Fresenius; travel funding from Merck, Grifols, and Bayer; and serves on advisory boards for Roche, Biogen, and Alexion. P.K. has received non-financial scientific grants from Miltenyi Biotec GmbH and the CECAD, University of Cologne, outside the submitted work. O.A.C. has received research grants from, is an advisor to, or received lecture honoraria from Actelion, Allecra, Amplyx, Astellas, Basilea, Biosys, Cidara, Da Volterra, Entasis, F2G, Gilead, Grupo Biotoscana, IQVIA, Janssen, Matinas, Medicines Company, MedPace, Melinta, Menarini, Merck/MSD, Mylan, Nabriva, Noxxon, Octapharma, Paratek, Pfizer, PSI, Roche Diagnostics, Scynexis, and Shionogi.
Figures


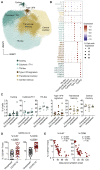

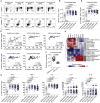

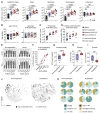
References
-
- Anft M., Paniskaki K., Blazquez-Navarro A., Doevelaar A.A.N., Seibert F., Hoelzer B., Skrzypczyk S., Kohut E., Kurek J., Zapka J., et al. COVID-19 progression is potentially driven by T cell immunopathogenesis. medRxiv. 2020 2020.2004.2028.20083089.
-
- Bacher P., Scheffold A. Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A. 2013;83:692–701. - PubMed
-
- Bacher P., Scheffold A. New technologies for monitoring human antigen-specific T cells and regulatory T cells by flow-cytometry. Curr. Opin. Pharmacol. 2015;23:17–24. - PubMed
-
- Bacher P., Schink C., Teutschbein J., Kniemeyer O., Assenmacher M., Brakhage A.A., Scheffold A. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J. Immunol. 2013;190:3967–3976. - PubMed
-
- Bacher P., Heinrich F., Stervbo U., Nienen M., Vahldieck M., Iwert C., Vogt K., Kollet J., Babel N., Sawitzki B., et al. Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell. 2016;167:1067–1078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

