Yes, we can use it: a formal test on the accuracy of low-pass nanopore long-read sequencing for mitophylogenomics and barcoding research using the Caribbean spiny lobster Panulirus argus
- PMID: 33297960
- PMCID: PMC7726883
- DOI: 10.1186/s12864-020-07292-5
Yes, we can use it: a formal test on the accuracy of low-pass nanopore long-read sequencing for mitophylogenomics and barcoding research using the Caribbean spiny lobster Panulirus argus
Abstract
Background: Whole mitogenomes or short fragments (i.e., 300-700 bp of the cox1 gene) are the markers of choice for revealing within- and among-species genealogies. Protocols for sequencing and assembling mitogenomes include 'primer walking' or 'long PCR' followed by Sanger sequencing or Illumina short-read low-coverage whole genome (LC-WGS) sequencing with or without prior enrichment of mitochondrial DNA. The aforementioned strategies assemble complete and accurate mitochondrial genomes but are time consuming and/or expensive. In this study, I first tested whether mitogenomes can be sequenced from long-read nanopore sequencing data exclusively. Second, I explored the accuracy of the long-read assembled genomes by comparing them to a 'gold' standard reference mitogenome retrieved from the same individual using Illumina sequencing. Third and lastly, I tested if the long-read assemblies are useful for mitophylogenomics and barcoding research. To accomplish these goals, I used the Caribbean spiny lobster Panulirus argus, an ecologically relevant species in shallow water coral reefs and target of the most lucrative fishery in the greater Caribbean region.
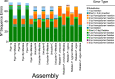
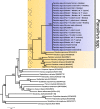
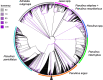
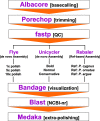
Results: LC-WGS using a MinION ONT device and various de-novo and reference-based assembly pipelines retrieved a complete and highly accurate mitogenome for the Caribbean spiny lobster Panulirus argus. Discordance between each of the long-read assemblies and the reference mitogenome was mostly due to indels at the flanks of homopolymer regions. Although not 'perfect', phylogenetic analyses using entire mitogenomes or a fragment of the cox1 gene demonstrated that mitogenomes assembled using long reads reliably identify the sequenced specimen as belonging to P. argus and distinguish it from other related species in the same genus, family, and superorder.
Conclusions: This study serves as a proof-of-concept for the future implementation of in-situ surveillance protocols using the MinION to detect mislabeling in P. argus across its supply chain. Mislabeling detection will improve fishery management in this overexploited lobster. This study will additionally aid in decreasing costs for exploring meta-population connectivity in the Caribbean spiny lobster and will aid with the transfer of genomics technology to low-income countries.
Keywords: Crayfish; Lobster; Long-read sequencing; Nanopore.
Conflict of interest statement
The author declares no competing interests.
Figures


References
-
- John U, Lu Y, Wohlrab S, Groth M, Janouškovec J, Kohli GS, Mark FC, Bickmeyer U, Farhat S, Felder M, Frickenhaus S. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Science Adv. 2019;5(4):eaav1110. doi: 10.1126/sciadv.aav1110. - DOI - PMC - PubMed
-
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10(1):50. doi: 10.1186/1471-2148-10-50. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

