A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition
- PMID: 33298160
- PMCID: PMC7724794
- DOI: 10.1186/s13073-020-00814-6
A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition
Abstract
Background: Antibiotic-resistant Klebsiella pneumoniae are a major cause of hospital- and community-acquired infections, including sepsis, liver abscess, and pneumonia, driven mainly by the emergence of successful high-risk clonal lineages. The K. pneumoniae sequence type (ST) 307 lineage has appeared in several different parts of the world after first being described in Europe in 2008. From June to October 2019, we recorded an outbreak of an extensively drug-resistant ST307 lineage in four medical facilities in north-eastern Germany.
Methods: Here, we investigated these isolates and those from subsequent cases in the same facilities. We performed whole-genome sequencing to study phylogenetics, microevolution, and plasmid transmission, as well as phenotypic experiments including growth curves, hypermucoviscosity, siderophore secretion, biofilm formation, desiccation resilience, serum survival, and heavy metal resistance for an in-depth characterization of this outbreak clone.
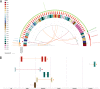
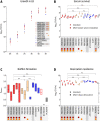
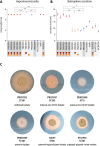
Results: Phylogenetics suggest a homogenous phylogram with several sub-clades containing either isolates from only one patient or isolates originating from different patients, suggesting inter-patient transmission. We identified three large resistance plasmids, carrying either NDM-1, CTX-M-15, or OXA-48, which K. pneumoniae ST307 likely donated to other K. pneumoniae isolates of different STs and even other bacterial species (e.g., Enterobacter cloacae) within the clinical settings. Several chromosomally and plasmid-encoded, hypervirulence-associated virulence factors (e.g., yersiniabactin, metabolite transporter, aerobactin, and heavy metal resistance genes) were identified in addition. While growth, biofilm formation, desiccation resilience, serum survival, and heavy metal resistance were comparable to several control strains, results from siderophore secretion and hypermucoviscosity experiments revealed superiority of the ST307 clone, similar to an archetypical, hypervirulent K. pneumoniae strain (hvKP1).
Conclusions: The combination of extensive drug resistance and virulence, partly conferred through a "mosaic" plasmid carrying both antibiotic resistance and hypervirulence-associated features, demonstrates serious public health implications.
Keywords: Hypervirulence; Outbreak; Plasmid transmission; XDR Klebsiella pneumoniae; “Mosaic” plasmid.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

