The Clathrin adaptor AP-1 and Stratum act in parallel pathways to control Notch activation in Drosophila sensory organ precursors cells
- PMID: 33298463
- PMCID: PMC7823167
- DOI: 10.1242/dev.191437
The Clathrin adaptor AP-1 and Stratum act in parallel pathways to control Notch activation in Drosophila sensory organ precursors cells
Abstract
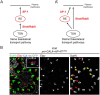
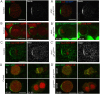
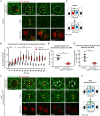
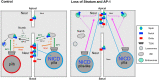
Drosophila sensory organ precursors divide asymmetrically to generate pIIa/pIIb cells, the identity of which relies on activation of Notch at cytokinesis. Although Notch is present apically and basally relative to the midbody at the pIIa-pIIb interface, the basal pool of Notch is reported to be the main contributor for Notch activation in the pIIa cell. Intra-lineage signalling requires appropriate apico-basal targeting of Notch, its ligand Delta and its trafficking partner Sanpodo. We have previously reported that AP-1 and Stratum regulate the trafficking of Notch and Sanpodo from the trans-Golgi network to the basolateral membrane. Loss of AP-1 or Stratum caused mild Notch gain-of-function phenotypes. Here, we report that their concomitant loss results in a penetrant Notch gain-of-function phenotype, indicating that they control parallel pathways. Although unequal partitioning of cell fate determinants and cell polarity were unaffected, we observed increased amounts of signalling-competent Notch as well as Delta and Sanpodo at the apical pIIa-pIIb interface, at the expense of the basal pool of Notch. We propose that AP-1 and Stratum operate in parallel pathways to localize Notch and control where receptor activation takes place.
Keywords: AP-1; Asymmetric cell division; Intracellular trafficking; Notch signalling; Stratum.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Aerts S., Quan X.-J., Claeys A., Naval Sanchez M., Tate P., Yan J. and Hassan B. A. (2010). Robust target gene discovery through transcriptome perturbations and genome-wide enhancer predictions in Drosophila uncovers a regulatory basis for sensory specification. PLoS Biol. 8, e1000435 10.1371/journal.pbio.1000435 - DOI - PMC - PubMed
-
- Bellaïche Y., Radovic A., Woods D. F., Hough C. D., Parmentier M.-L., O'Kane C. J., Bryant P. J. and Schweisguth F. (2001). The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell 106, 355-366. 10.1016/S0092-8674(01)00444-5 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

