Detection of Five mcr-9-Carrying Enterobacterales Isolates in Four Czech Hospitals
- PMID: 33298573
- PMCID: PMC7729258
- DOI: 10.1128/mSphere.01008-20
Detection of Five mcr-9-Carrying Enterobacterales Isolates in Four Czech Hospitals
Abstract
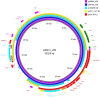
The aim of this study was to report the characterization of the first mcr-positive Enterobacterales isolated from Czech hospitals. In 2019, one Citrobacter freundii and four Enterobacter isolates were recovered from Czech hospitals. The production of carbapenemases was examined by a matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) imipenem hydrolysis assay. Additionally, bacteria were screened for the presence of carbapenemase-encoding genes and plasmid-mediated colistin resistance genes by PCR. To define the genetic units carrying mcr genes, the genomic DNAs of mcr-carrying clinical isolates were sequenced on the PacBio Sequel I platform. Results showed that all isolates carried blaVIM- and mcr-like genes. Analysis of whole-genome sequencing (WGS) data revealed that all isolates carried mcr-9-like alleles. Furthermore, the three sequence type 106 (ST106) Enterobacter hormaechei isolates harbored the blaVIM-1 gene, while the ST764 E. hormaechei and ST95 C. freundii included blaVIM-4 Analysis of plasmid sequences showed that, in all isolates, mcr-9 was carried on IncHI2 plasmids. Additionally, at least one multidrug resistance (MDR) region was identified in each mcr-9-carrying IncHI2 plasmid. The blaVIM-4 gene was found in the MDR regions of p48880_MCR_VIM and p51929_MCR_VIM. In the three remaining isolates, blaVIM-1 was localized on plasmids (∼55 kb) exhibiting repA-like sequences 99% identical to the respective gene of pKPC-CAV1193. In conclusion, to the best of our knowledge, these 5 isolates were the first mcr-9-positive bacteria of clinical origin identified in the Czech Republic. Additionally, the carriage of the blaVIM-1 on pKPC-CAV1193-like plasmids is described for the first time. Thus, our findings underline the ongoing evolution of mobile elements implicated in the dissemination of clinically important resistance determinants.IMPORTANCE Infections caused by carbapenemase-producing bacteria have led to the revival of polymyxins as the "last-resort" antibiotic. Since 2016, several reports describing the presence of plasmid-mediated colistin resistance genes, mcr, in different host species and geographic areas were published. Here, we report the first detection of Enterobacterales carrying mcr-9-like alleles isolated from Czech hospitals in 2019. Furthermore, the three ST106 Enterobacter hormaechei isolates harbored blaVIM-1, while the ST764 E. hormaechei and ST95 Citrobacter freundii isolates included blaVIM-4 Analysis of WGS data showed that, in all isolates, mcr-9 was carried on IncHI2 plasmids. blaVIM-4 was found in the MDR regions of IncHI2 plasmids, while blaVIM-1 was localized on pKPC-CAV1193-like plasmids, described here for the first time. These findings underline the ongoing evolution of mobile elements implicated in dissemination of clinically important resistance determinants. Thus, WGS characterization of MDR bacteria is crucial to unravel the mechanisms involved in dissemination of resistance mechanisms.
Keywords: Citrobacter freundii; Enterobacter cloacae; IncHI2; MCR-9; VIM-4.
Copyright © 2020 Bitar et al.
Figures
References
-
- Yilmaz GR, Dizbay M, Guven T, Pullukcu H, Tasbakan M, Guzel OT, Tekce YT, Ozden M, Turhan O, Guner R, Cag Y, Bozkurt F, Karadag FY, Kartal ED, Gozel G, Bulut C, Erdinc S, Keske S, Acikgoz ZC, Tasyaran MA. 2016. Risk factors for infection with colistin-resistant gram-negative microorganisms: a multicenter study. Ann Saudi Med 36:216–222. doi:10.5144/0256-4947.2016.216. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous