Xpert MTB/XDR: a 10-Color Reflex Assay Suitable for Point-of-Care Settings To Detect Isoniazid, Fluoroquinolone, and Second-Line-Injectable-Drug Resistance Directly from Mycobacterium tuberculosis-Positive Sputum
- PMID: 33298611
- PMCID: PMC8106700
- DOI: 10.1128/JCM.02314-20
Xpert MTB/XDR: a 10-Color Reflex Assay Suitable for Point-of-Care Settings To Detect Isoniazid, Fluoroquinolone, and Second-Line-Injectable-Drug Resistance Directly from Mycobacterium tuberculosis-Positive Sputum
Abstract
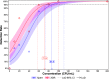
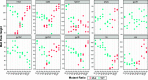
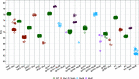
We describe the design, development, analytical performance, and a limited clinical evaluation of the 10-color Xpert MTB/XDR assay (CE-IVD only, not for sale in the United States). This assay is intended as a reflex test to detect resistance to isoniazid (INH), fluoroquinolones (FLQ), ethionamide (ETH), and second-line injectable drugs (SLIDs) in unprocessed sputum samples and concentrated sputum sediments which are positive for Mycobacterium tuberculosis The Xpert MTB/XDR assay simultaneously amplifies eight genes and promoter regions in M. tuberculosis and analyzes melting temperatures (Tm s) using sloppy molecular beacon (SMB) probes to identify mutations associated with INH, FLQ, ETH, and SLID resistance. Results can be obtained in under 90 min using 10-color GeneXpert modules. The assay can differentiate low- versus high-level resistance to INH and FLQ as well as cross-resistance versus individual resistance to SLIDs by identifying mutation-specific Tm s or Tm patterns generated by the SMB probes. The assay has a limit of detection comparable to that of the Xpert MTB/RIF assay and successfully detected 16 clinically significant mutations in a challenge set of clinical isolate DNA. In a clinical study performed at two sites with 100 sputum and 214 clinical isolates, the assay showed a sensitivity of 94% to 100% and a specificity of 100% for all drugs except for ETH compared to that of sequencing. The sensitivity and specificity were in the same ranges as those of phenotypic drug-susceptibility testing. Used in combination with a primary tuberculosis diagnostic test, this assay should expand the capacity for detection of drug-resistant tuberculosis near the point of care.
Keywords: 10-color; GeneXpert; Mycobacterium tuberculosis; TB; XDR; drug resistance; rapid diagnostics.
Copyright © 2021 American Society for Microbiology.
Figures
References
-
- World Health Organization. 2019. Global tuberculosis report 2019. World Health Organization, Geneva, Switzerland.
-
- Georghiou SB, Schumacher SG, Rodwell TC, Colman RE, Miotto P, Gilpin C, Ismail N, Rodrigues C, Warren R, Weyer K, Zignol M, Arafah S, Cirillo DM, Denkinger CM. 2019. Guidance for studies evaluating the accuracy of rapid tuberculosis drug-susceptibility tests. J Infect Dis 220:S126–S135. doi: 10.1093/infdis/jiz106. - DOI - PubMed
-
- Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. - DOI - PMC - PubMed
-
- Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, Hall SL, Chakravorty S, Cirillo DM, Tukvadze N, Bablishvili N, Stevens W, Scott L, Rodrigues C, Kazi MI, Joloba M, Nakiyingi L, Nicol MP, Ghebrekristos Y, Anyango I, Murithi W, Dietze R, Lyrio Peres R, Skrahina A, Auchynka V, Chopra KK, Hanif M, Liu X, Yuan X, Boehme CC, Ellner JJ, Denkinger CM. 2018. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis 18:76–84. doi: 10.1016/S1473-3099(17)30691-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

