Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection
- PMID: 33298930
- PMCID: PMC7725958
- DOI: 10.1038/s41467-020-20139-7
Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection
Abstract
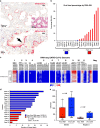
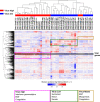
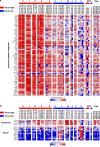
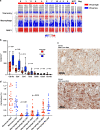
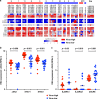
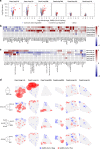
The relationship of SARS-CoV-2 pulmonary infection and severity of disease is not fully understood. Here we show analysis of autopsy specimens from 24 patients who succumbed to SARS-CoV-2 infection using a combination of different RNA and protein analytical platforms to characterize inter-patient and intra-patient heterogeneity of pulmonary virus infection. There is a spectrum of high and low virus cases associated with duration of disease. High viral cases have high activation of interferon pathway genes and a predominant M1-like macrophage infiltrate. Low viral cases are more heterogeneous likely reflecting inherent patient differences in the evolution of host response, but there is consistent indication of pulmonary epithelial cell recovery based on napsin A immunohistochemistry and RNA expression of surfactant and mucin genes. Using a digital spatial profiling platform, we find the virus corresponds to distinct spatial expression of interferon response genes demonstrating the intra-pulmonary heterogeneity of SARS-CoV-2 infection.
Conflict of interest statement
D.T.T. declares having received a speaker fee for participation in a conference supported by NanoString, Inc. about this work. D.T.T. declares receiving consulting fees from Pfizer, Third Rock Ventures, Merrimack Pharmaceuticals, Ventana Roche, Foundation Medicine, Inc., and EMD Millipore Sigma, which are not related to this work. D.T.T. declares that he is a founder and has equity in PanTher Therapeutics and TellBio, Inc., which is not related to this work. D.T.T. and B.D.G. declare they are co-founders and own equity in ROME Therapeutics, which is not related to this work. N.D., A.N., A.S.K., V.D., M.N.R. and D.T.T. declare they are supported by ACD-Biotechne. Robert Monroe declares that he is an employee of ACD-Biotechne. Sarah E Warren, Patrick Danaher, Jason W. Reeves, Jingjing Gong, Erroll H Rueckert declare that they are employees of NanoString, Inc. The other authors declare no competing interests.
Figures

Update of
-
Temporal and Spatial Heterogeneity of Host Response to SARS-CoV-2 Pulmonary Infection.medRxiv [Preprint]. 2020 Aug 2:2020.07.30.20165241. doi: 10.1101/2020.07.30.20165241. medRxiv. 2020. Update in: Nat Commun. 2020 Dec 9;11(1):6319. doi: 10.1038/s41467-020-20139-7. PMID: 32766600 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

