Missense variant contribution to USP9X-female syndrome
- PMID: 33298948
- PMCID: PMC7725775
- DOI: 10.1038/s41525-020-00162-9
Missense variant contribution to USP9X-female syndrome
Abstract
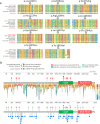
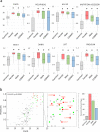
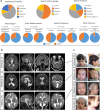
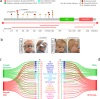
USP9X is an X-chromosome gene that escapes X-inactivation. Loss or compromised function of USP9X leads to neurodevelopmental disorders in males and females. While males are impacted primarily by hemizygous partial loss-of-function missense variants, in females de novo heterozygous complete loss-of-function mutations predominate, and give rise to the clinically recognisable USP9X-female syndrome. Here we provide evidence of the contribution of USP9X missense and small in-frame deletion variants in USP9X-female syndrome also. We scrutinise the pathogenicity of eleven such variants, ten of which were novel. Combined application of variant prediction algorithms, protein structure modelling, and assessment under clinically relevant guidelines universally support their pathogenicity. The core phenotype of this cohort overlapped with previous descriptions of USP9X-female syndrome, but exposed heightened variability. Aggregate phenotypic information of 35 currently known females with predicted pathogenic variation in USP9X reaffirms the clinically recognisable USP9X-female syndrome, and highlights major differences when compared to USP9X-male associated neurodevelopmental disorders.
Conflict of interest statement
L.A. P.-J. is scientific advisor and founding partner of qGenomics Laboratory. All other authors have no conflicts of interest or financial disclosures to declare.
Figures