This is a preprint.
Designed proteins assemble antibodies into modular nanocages
- PMID: 33299994
- PMCID: PMC7724662
- DOI: 10.1101/2020.12.01.406611
Designed proteins assemble antibodies into modular nanocages
Update in
-
Designed proteins assemble antibodies into modular nanocages.Science. 2021 Apr 2;372(6537):eabd9994. doi: 10.1126/science.abd9994. Science. 2021. PMID: 33795432 Free PMC article.
Abstract
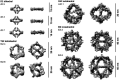
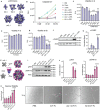
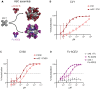
Antibodies are widely used in biology and medicine, and there has been considerable interest in multivalent antibody formats to increase binding avidity and enhance signaling pathway agonism. However, there are currently no general approaches for forming precisely oriented antibody assemblies with controlled valency. We describe the computational design of two-component nanocages that overcome this limitation by uniting form and function. One structural component is any antibody or Fc fusion and the second is a designed Fc-binding homo-oligomer that drives nanocage assembly. Structures of 8 antibody nanocages determined by electron microscopy spanning dihedral, tetrahedral, octahedral, and icosahedral architectures with 2, 6, 12, and 30 antibodies per nanocage match the corresponding computational models. Antibody nanocages targeting cell-surface receptors enhance signaling compared to free antibodies or Fc-fusions in DR5-mediated apoptosis, Tie2-mediated angiogenesis, CD40 activation, and T cell proliferation; nanocage assembly also increases SARS-CoV-2 pseudovirus neutralization by α-SARS-CoV-2 monoclonal antibodies and Fc-ACE2 fusion proteins. We anticipate that the ability to assemble arbitrary antibodies without need for covalent modification into highly ordered assemblies with different geometries and valencies will have broad impact in biology and medicine.
Conflict of interest statement
Competing Interests Provisional patents have been filed on the AbC-forming designs, α-DR5 AbCs, A1F-Fc AbCs, α-CD40 AbCs, and α-CoV-2 S AbCs. A provisional patent application (U.S. Provisional Application No. 63/016268) has been filed on the SARS-CoV-2 specific monoclonal antibodies discussed here. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. The other authors declare no competing interests.
Figures



References
-
- Cuesta A. M., Sainz-Pastor N., Bonet J., Oliva B., Alvarez-Vallina L., Multivalent antibodies: when design surpasses evolution. Trends Biotechnol. 28, 355–362 (2010). - PubMed
-
- Nuñez-Prado N., Compte M., Harwood S., Álvarez-Méndez A., Lykkemark S., Sanz L., Álvarez-Vallina L., The coming of age of engineered multivalent antibodies. Drug Discov. Today. 20, 588–594 (2015). - PubMed
-
- Laursen N. S., Friesen R. H. E., Zhu X., Jongeneelen M., Blokland S., Vermond J., van Eijgen A., Tang C., van Diepen H., Obmolova G., van der Neut Kolfschoten M., Zuijdgeest D., Straetemans R., Hoffman R. M. B., Nieusma T., Pallesen J., Turner H. L., Bernard S. M., Ward A. B., Luo J., Poon L. L. M., Tretiakova A. P., Wilson J. M., Limberis M. P., Vogels R., Brandenburg B., Kolkman J. A., Wilson I. A., Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 362, 598–602 (2018). - PMC - PubMed
-
- Seifert O., Plappert A., Fellermeier S., Siegemund M., Pfizenmaier K., Kontermann R. E., Tetravalent antibody-scTRAIL fusion proteins with improved properties. Mol. Cancer Ther. 13, 101–111 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
