Aberrant mitochondrial morphology and function associated with impaired mitophagy and DNM1L-MAPK/ERK signaling are found in aged mutant Parkinsonian LRRK2R1441G mice
- PMID: 33300446
- PMCID: PMC8526027
- DOI: 10.1080/15548627.2020.1850008
Aberrant mitochondrial morphology and function associated with impaired mitophagy and DNM1L-MAPK/ERK signaling are found in aged mutant Parkinsonian LRRK2R1441G mice
Abstract
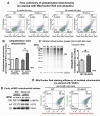
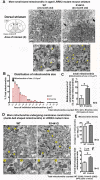
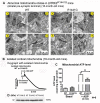
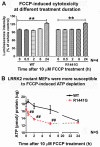
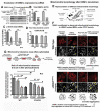
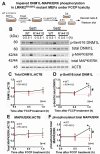
Mitochondrial dysfunction causes energy deficiency and nigrostriatal neurodegeneration which is integral to the pathogenesis of Parkinson disease (PD). Clearance of defective mitochondria involves fission and ubiquitin-dependent degradation via mitophagy to maintain energy homeostasis. We hypothesize that LRRK2 (leucine-rich repeat kinase 2) mutation disrupts mitochondrial turnover causing accumulation of defective mitochondria in aging brain. We found more ubiquitinated mitochondria with aberrant morphology associated with impaired function in aged (but not young) LRRK2R1441G knockin mutant mouse striatum compared to wild-type (WT) controls. LRRK2R1441G mutant mouse embryonic fibroblasts (MEFs) exhibited reduced MAP1LC3/LC3 activation indicating impaired macroautophagy/autophagy. Mutant MEFs under FCCP-induced (mitochondrial uncoupler) stress showed increased LC3-aggregates demonstrating impaired mitophagy. Using a novel flow cytometry assay to quantify mitophagic rates in MEFs expressing photoactivatable mito-PAmCherry, we found significantly slower mitochondria clearance in mutant cells. Specific LRRK2 kinase inhibition using GNE-7915 did not alleviate impaired mitochondrial clearance suggesting a lack of direct relationship to increased kinase activity alone. DNM1L/Drp1 knockdown in MEFs slowed mitochondrial clearance indicating that DNM1L is a prerequisite for mitophagy. DNM1L knockdown in slowing mitochondrial clearance was less pronounced in mutant MEFs, indicating preexisting impaired DNM1L activation. DNM1L knockdown disrupted mitochondrial network which was more evident in mutant MEFs. DNM1L-Ser616 and MAPK/ERK phosphorylation which mediate mitochondrial fission and downstream mitophagic processes was apparent in WT using FCCP-induced stress but not mutant MEFs, despite similar total MAPK/ERK and DNM1L levels. In conclusion, aberrant mitochondria morphology and dysfunction associated with impaired mitophagy and DNM1L-MAPK/ERK signaling are found in mutant LRRK2 MEFs and mouse brain.Abbreviations: ATP: adenosine triphosphate; BAX: BCL2-associated X protein; CDK1: cyclin-dependent kinase 1; CDK5: cyclin-dependent kinase 5; CQ: chloroquine; CSF: cerebrospinal fluid; DNM1L/DRP1: dynamin 1-like; ELISA: enzyme-linked immunosorbent assay; FACS: fluorescence-activated cell sorting; FCCP: carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; LAMP2A: lysosomal-associated membrane protein 2A; LRRK2: leucine-rich repeat kinase 2; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MAPK1/ERK2: mitogen-activated protein kinase 1; MEF: mouse embryonic fibroblast; MFN1: mitofusin 1; MMP: mitochondrial membrane potential; PAmCherry: photoactivatable-mCherry; PD: Parkinson disease; PINK1: PTEN induced putative kinase 1; PRKN/PARKIN: parkin RBR E3 ubiquitin protein ligase; RAB10: RAB10, member RAS oncogene family; RAF: v-raf-leukemia oncogene; SNCA: synuclein, alpha; TEM: transmission electron microscopy; VDAC: voltage-dependent anion channel; WT: wild type; SQSTM1/p62: sequestosome 1.
Keywords: Aging; Dnm1l/DRP1; SQSTM1/p62; knockin mice; macroautophagy; mitochondria dysfunction; mitochondrial fission; mitophagy; parkinson disease; ubiquitination.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
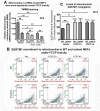
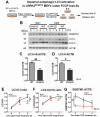
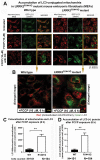
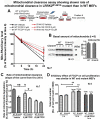
References
-
- Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol. 2008;7(1):97–109. - PubMed
-
- Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci. 2006;7(3):207–219. - PubMed
-
- Sprenger HG, The LT. Good and the bad of mitochondrial breakups. Trends Cell Biol. 2019;29(11):888–900. - PubMed
-
- Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19(2):93–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
