Prioritizing genes for systematic variant effect mapping
- PMID: 33300982
- PMCID: PMC8016487
- DOI: 10.1093/bioinformatics/btaa1008
Prioritizing genes for systematic variant effect mapping
Abstract
Motivation: When rare missense variants are clinically interpreted as to their pathogenicity, most are classified as variants of uncertain significance (VUS). Although functional assays can provide strong evidence for variant classification, such results are generally unavailable. Multiplexed assays of variant effect can generate experimental 'variant effect maps' that score nearly all possible missense variants in selected protein targets for their impact on protein function. However, these efforts have not always prioritized proteins for which variant effect maps would have the greatest impact on clinical variant interpretation.
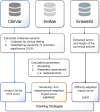
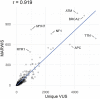
Results: Here, we mined databases of clinically interpreted variants and applied three strategies, each building on the previous, to prioritize genes for systematic functional testing of missense variation. The strategies ranked genes (i) by the number of unique missense VUS that had been reported to ClinVar; (ii) by movability- and reappearance-weighted impact scores, to give extra weight to reappearing, movable VUS and (iii) by difficulty-adjusted impact scores, to account for the more resource-intensive nature of generating variant effect maps for longer genes. Our results could be used to guide systematic functional testing of missense variation toward greater impact on clinical variant interpretation.
Availability and implementation: Source code available at: https://github.com/rothlab/mave-gene-prioritization.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures

References
-
- Arad M. et al. (2005) Gene mutations in apical hypertrophic cardiomyopathy. Circulation, 112, 2805–2811. - PubMed

