Synthetic Tuning of Domain Stoichiometry in Nanobody-Enzyme Megamolecules
- PMID: 33301672
- PMCID: PMC8109025
- DOI: 10.1021/acs.bioconjchem.0c00578
Synthetic Tuning of Domain Stoichiometry in Nanobody-Enzyme Megamolecules
Abstract
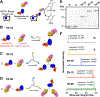
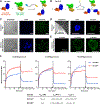
This paper presents a method to synthetically tune atomically precise megamolecule nanobody-enzyme conjugates for prodrug cancer therapy. Previous efforts to create heterobifunctional protein conjugates suffered from heterogeneity in domain stoichiometry, which in part led to the failure of antibody-enzyme conjugates in clinical trials. We used the megamolecule approach to synthesize anti-HER2 nanobody-cytosine deaminase conjugates with tunable numbers of nanobody and enzyme domains in a single, covalent molecule. Linking two nanobody domains to one enzyme domain improved avidity to a human cancer cell line by 4-fold but did not increase cytotoxicity significantly due to lowered enzyme activity. In contrast, a megamolecule composed of one nanobody and two enzyme domains resulted in an 8-fold improvement in the catalytic efficiency and increased the cytotoxic effect by over 5-fold in spheroid culture, indicating that the multimeric structure allowed for an increase in local drug activation. Our work demonstrates that the megamolecule strategy can be used to study structure-function relationships of protein conjugate therapeutics with synthetic control of protein domain stoichiometry.
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures



Similar articles
-
Dual-Targeting of the HER2 Cancer Receptor with an Antibody-Directed Enzyme and a Nanobody-Guided MMAE Prodrug Scaffold.Chembiochem. 2024 Sep 16;25(18):e202400437. doi: 10.1002/cbic.202400437. Epub 2024 Aug 20. Chembiochem. 2024. PMID: 38945824
-
Activation of prodrugs by antibody-enzyme conjugates: a new approach to cancer therapy.FASEB J. 1990 Feb 1;4(2):188-93. doi: 10.1096/fasebj.4.2.2404820. FASEB J. 1990. PMID: 2404820 Review.
-
Modular design of nanobody-drug conjugates for targeted-delivery of platinum anticancer drugs with an MRI contrast agent.Chem Commun (Camb). 2019 Apr 25;55(35):5175-5178. doi: 10.1039/c9cc01391a. Chem Commun (Camb). 2019. PMID: 30984937
-
Conjugation of oxaliplatin with PEGylated-nanobody for enhancing tumor targeting and prolonging circulation.J Inorg Biochem. 2021 Oct;223:111553. doi: 10.1016/j.jinorgbio.2021.111553. Epub 2021 Jul 24. J Inorg Biochem. 2021. PMID: 34340059
-
Selective activation of anticancer prodrugs by monoclonal antibody-enzyme conjugates.Methods Find Exp Clin Pharmacol. 1994 Sep;16(7):505-12. Methods Find Exp Clin Pharmacol. 1994. PMID: 7885077 Review.
Cited by
-
Blueprints for Better Drugs: The Structural Revolution in Nanomedicine.ACS Nano. 2025 May 27;19(20):18889-18901. doi: 10.1021/acsnano.5c06380. Epub 2025 May 13. ACS Nano. 2025. PMID: 40359339
References
-
- Sharma SK, and Bagshawe KD (2017) Antibody Directed Enzyme Prodrug Therapy (ADEPT): Trials and tribulations. Adv. Drug Delivery Rev 118, 2–7. - PubMed
-
- Bagshawe KD (2006) Antibody-directed enzyme prodrug therapy (ADEPT) for cancer. Expert Rev. Anticancer Ther 6, 1421–1431. - PubMed
-
- Senter PD, and Springer CJ (2001) Selective activation of anticancer prodrugs by monoclonal antibody–enzyme conjugates. Adv. Drug Delivery Rev 53, 247–264. - PubMed
-
- Melton RG, and Sherwood RF (1996) antibody–enzyme Conjugates for Cancer Therapy. J. Natl. Cancer Inst 88, 153–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

