Role of autophagy in intervertebral disc and cartilage function: implications in health and disease
- PMID: 33301899
- PMCID: PMC8180533
- DOI: 10.1016/j.matbio.2020.12.002
Role of autophagy in intervertebral disc and cartilage function: implications in health and disease
Abstract
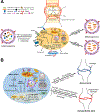
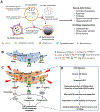
The intervertebral disc and cartilage are specialized, extracellular matrix-rich tissues critical for absorbing mechanical loads, providing flexibility to the joints, and longitudinal growth in the case of growth plate cartilage. Specialized niche conditions in these tissues, such as hypoxia, are critical in regulating cellular activities including autophagy, a lysosomal degradation pathway that promotes cell survival. Mounting evidence suggests that dysregulation of autophagic pathways underscores many skeletal pathologies affecting the spinal column, articular and growth plate cartilages. Many lysosomal storage disorders characterized by the accumulation of partially degraded glycosaminoglycans (GAGs) due to the lysosomal dysfunction thus affect skeletal tissues and result in altered ECM structure. Likewise, pathologies that arise from mutations in genes encoding ECM proteins and ECM processing, folding, and post-translational modifications, result in accumulation of misfolded proteins in the ER, ER stress and autophagy dysregulation. These conditions evidence reduced secretion of ECM proteins and/or increased secretion of mutant proteins, thereby impairing matrix quality and the integrity of affected skeletal tissues and causing a lack of growth and degeneration. In this review, we discuss the role of autophagy and mechanisms of its regulation in the intervertebral disc and cartilages, as well as how dysregulation of autophagic pathways affects these skeletal tissues.
Keywords: Articular cartilage; Autophagy; Chondrodysplasia; Disc degeneration; ECM mutations; ER stress; GAG storage disorders; Growth plate; Intervertebral disc; Lysosomal storage disorders; Osteoarthritis; Unfolded protein response.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflicts of interest.
Figures
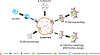
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

