Aberrant Activity of Histone-Lysine N-Methyltransferase 2 (KMT2) Complexes in Oncogenesis
- PMID: 33302406
- PMCID: PMC7762615
- DOI: 10.3390/ijms21249340
Aberrant Activity of Histone-Lysine N-Methyltransferase 2 (KMT2) Complexes in Oncogenesis
Abstract
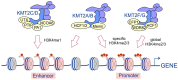
KMT2 (histone-lysine N-methyltransferase subclass 2) complexes methylate lysine 4 on the histone H3 tail at gene promoters and gene enhancers and, thus, control the process of gene transcription. These complexes not only play an essential role in normal development but have also been described as involved in the aberrant growth of tissues. KMT2 mutations resulting from the rearrangements of the KMT2A (MLL1) gene at 11q23 are associated with pediatric mixed-lineage leukemias, and recent studies demonstrate that KMT2 genes are frequently mutated in many types of human cancers. Moreover, other components of the KMT2 complexes have been reported to contribute to oncogenesis. This review summarizes the recent advances in our knowledge of the role of KMT2 complexes in cell transformation. In addition, it discusses the therapeutic targeting of different components of the KMT2 complexes.
Keywords: COMPASS; COMPASS-like; H3K4 methylation; cancer; chromatin; epigenetics; histone–lysine N-methyltransferase 2; oncogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Histone-lysine N-methyltransferase 2 (KMT2) complexes - a new perspective.Mutat Res Rev Mutat Res. 2022 Jul-Dec;790:108443. doi: 10.1016/j.mrrev.2022.108443. Epub 2022 Sep 22. Mutat Res Rev Mutat Res. 2022. PMID: 36154872 Review.
-
Transcriptional regulation by the KMT2 histone H3K4 methyltransferases.Biochim Biophys Acta Gene Regul Mech. 2020 Jul;1863(7):194545. doi: 10.1016/j.bbagrm.2020.194545. Epub 2020 Mar 16. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32194213 Review.
-
Hijacked in cancer: the KMT2 (MLL) family of methyltransferases.Nat Rev Cancer. 2015 Jun;15(6):334-46. doi: 10.1038/nrc3929. Nat Rev Cancer. 2015. PMID: 25998713 Free PMC article. Review.
-
Biochemical perspectives on targeting KMT2 methyltransferases in cancer.Trends Pharmacol Sci. 2021 Aug;42(8):688-699. doi: 10.1016/j.tips.2021.05.002. Epub 2021 May 30. Trends Pharmacol Sci. 2021. PMID: 34074527 Review.
-
COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer.Cancer Lett. 2019 Aug 28;458:56-65. doi: 10.1016/j.canlet.2019.05.024. Epub 2019 May 22. Cancer Lett. 2019. PMID: 31128216 Free PMC article. Review.
Cited by
-
KMT2A facilitates the epithelial-to-mesenchymal transition and the progression of ovarian cancer.Mol Cell Biochem. 2025 May;480(5):3001-3017. doi: 10.1007/s11010-024-05167-x. Epub 2024 Nov 26. Mol Cell Biochem. 2025. PMID: 39589456
-
The Ubiquitin-Conjugating Enzyme E2 O (UBE2O) and Its Therapeutic Potential in Human Leukemias and Solid Tumors.Cancers (Basel). 2024 Sep 3;16(17):3064. doi: 10.3390/cancers16173064. Cancers (Basel). 2024. PMID: 39272922 Free PMC article. Review.
-
Induction of senescence upon loss of the Ash2l core subunit of H3K4 methyltransferase complexes.Nucleic Acids Res. 2022 Aug 12;50(14):7889-7905. doi: 10.1093/nar/gkac591. Nucleic Acids Res. 2022. PMID: 35819198 Free PMC article.
-
Mutations in Epigenetic Regulation Genes in Gastric Cancer.Cancers (Basel). 2021 Sep 13;13(18):4586. doi: 10.3390/cancers13184586. Cancers (Basel). 2021. PMID: 34572812 Free PMC article.
-
KRAS Promotes GLI2-Dependent Transcription during Pancreatic Carcinogenesis.Cancer Res Commun. 2024 Jul 1;4(7):1677-1689. doi: 10.1158/2767-9764.CRC-23-0464. Cancer Res Commun. 2024. PMID: 38896052 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

