Metabolomic Profile of Personalized Donor Human Milk
- PMID: 33302441
- PMCID: PMC7763631
- DOI: 10.3390/molecules25245783
Metabolomic Profile of Personalized Donor Human Milk
Abstract
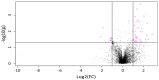
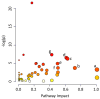
Human milk could be considered an active and complex mixture of beneficial bacteria and bioactive compounds. Since pasteurization drastically reduces the microbial content, we recently demonstrated that pasteurized donor human milk (DHM) could be inoculated with different percentages (10% and 30%) of mother's own milk (MOM) to restore the unique live microbiota, resulting in personalized milk (RM10 and RM30, respectively). Pasteurization affects not only the survival of the microbiota but also the concentration of proteins and metabolites, in this study, we performed a comparative metabolomic analysis of the RM10, RM30, MOM and DHM samples to evaluate the impact of microbial restoration on metabolite profiles, where metabolite profiles clustered into four well-defined groups. Comparative analyses of DHM and MOM metabolomes determined that over one thousand features were significantly different. In addition, significant changes in the metabolite concentrations were observed in MOM and RM30 samples after four hours of incubation, while the concentration of metabolites in DHM remained constant, indicating that these changes are related to the microbial expansion. In summary, our analyses indicate that the metabolite profiles of DHM are significantly different from that of MOM, and the profile of MOM may be partially restored in DHM through microbial expansion.
Keywords: metabolomics; microbiota; mother’s own milk.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Ziegler E.E. Protein and Energy Requirements in Infancy and Childhood. Volume 58. Karger Publishers; Basel, Switzerland: 2006. Growth of Breast-Fed and Formula-Fed Infants; pp. 51–63. - PubMed
-
- WHO [(accessed on 1 June 2020)];Exclusive Breastfeeding for Six Months Best for Babies Everywhere. Available online: https://www.who.int/maternal_child_adolescent/news_events/news/2011/20_0...
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

