Unique Mitochondrial Single Nucleotide Polymorphisms Demonstrate Resolution Potential to Discriminate Theileria parva Vaccine and Buffalo-Derived Strains
- PMID: 33302571
- PMCID: PMC7764068
- DOI: 10.3390/life10120334
Unique Mitochondrial Single Nucleotide Polymorphisms Demonstrate Resolution Potential to Discriminate Theileria parva Vaccine and Buffalo-Derived Strains
Abstract
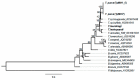
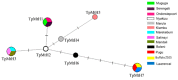
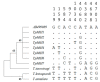
Distinct pathogenic and epidemiological features underlie different Theileria parva strains resulting in different clinical manifestations of East Coast Fever and Corridor Disease in susceptible cattle. Unclear delineation of these strains limits the control of these diseases in endemic areas. Hence, an accurate characterization of strains can improve the treatment and prevention approaches as well as investigate their origin. Here, we describe a set of single nucleotide polymorphisms (SNPs) based on 13 near-complete mitogenomes of T. parva strains originating from East and Southern Africa, including the live vaccine stock strains. We identified 11 SNPs that are non-preferentially distributed within the coding and non-coding regions, all of which are synonymous except for two within the cytochrome b gene of buffalo-derived strains. Our analysis ascertains haplotype-specific mutations that segregate the different vaccine and the buffalo-derived strains except T. parva-Muguga and Serengeti-transformed strains suggesting a shared lineage between the latter two vaccine strains. Phylogenetic analyses including the mitogenomes of other Theileria species: T. annulata, T. taurotragi, and T. lestoquardi, with the latter two sequenced in this study for the first time, were congruent with nuclear-encoded genes. Importantly, we describe seven T. parva haplotypes characterized by synonymous SNPs and parsimony-informative characters with the other three transforming species mitogenomes. We anticipate that tracking T. parva mitochondrial haplotypes from this study will provide insight into the parasite's epidemiological dynamics and underpin current control efforts.
Keywords: SNPs; Theileria parva; haplotypes; live vaccine; mitogenomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Norval R.A.I., Perry B.D. The Epidemiology of Theileriosis in Africa. Academic Press; London, UK: 1992. pp. 1–41.
Grants and funding
LinkOut - more resources
Full Text Sources

