Common Factors in Neurodegeneration: A Meta-Study Revealing Shared Patterns on a Multi-Omics Scale
- PMID: 33302607
- PMCID: PMC7764447
- DOI: 10.3390/cells9122642
Common Factors in Neurodegeneration: A Meta-Study Revealing Shared Patterns on a Multi-Omics Scale
Abstract
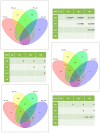
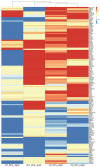
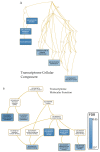
Neurodegenerative diseases such as Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and amyotrophic lateral sclerosis (ALS) are heterogeneous, progressive diseases with frequently overlapping symptoms characterized by a loss of neurons. Studies have suggested relations between neurodegenerative diseases for many years (e.g., regarding the aggregation of toxic proteins or triggering endogenous cell death pathways). We gathered publicly available genomic, transcriptomic, and proteomic data from 177 studies and more than one million patients to detect shared genetic patterns between the neurodegenerative diseases on three analyzed omics-layers. The results show a remarkably high number of shared differentially expressed genes between the transcriptomic and proteomic levels for all conditions, while showing a significant relation between genomic and proteomic data between AD and PD and AD and ALS. We identified a set of 139 genes being differentially expressed in several transcriptomic experiments of all four diseases. These 139 genes showed overrepresented gene ontology (GO) Terms involved in the development of neurodegeneration, such as response to heat and hypoxia, positive regulation of cytokines and angiogenesis, and RNA catabolic process. Furthermore, the four analyzed neurodegenerative diseases (NDDs) were clustered by their mean direction of regulation throughout all transcriptomic studies for this set of 139 genes, with the closest relation regarding this common gene set seen between AD and HD. GO-Term and pathway analysis of the proteomic overlap led to biological processes (BPs), related to protein folding and humoral immune response. Taken together, we could confirm the existence of many relations between Alzheimer's disease, Parkinson's disease, Huntington's disease, and amyotrophic lateral sclerosis on transcriptomic and proteomic levels by analyzing the pathways and GO-Terms arising in these intersections. The significance of the connection and the striking relation of the results to processes leading to neurodegeneration between the transcriptomic and proteomic data for all four analyzed neurodegenerative diseases showed that exploring many studies simultaneously, including multiple omics-layers of different neurodegenerative diseases simultaneously, holds new relevant insights that do not emerge from analyzing these data separately. Furthermore, the results shed light on processes like the humoral immune response that have previously been described only for certain diseases. Our data therefore suggest human patients with neurodegenerative diseases should be addressed as complex biological systems by integrating multiple underlying data sources.
Keywords: Alzheimer’s disease; Huntington’s disease; Parkinson’s disease; amyotrophic lateral sclerosis; multi-omics; neurodegeneration.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Xie T., Deng L., Mei P., Zhou Y., Wang B., Zhang J., Lin J., Wei Y., Zhang X., Xu R. A genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol. Aging. 2014;35:1778.e9–1778.e23. doi: 10.1016/j.neurobiolaging.2014.01.014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

