Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations
- PMID: 33302617
- PMCID: PMC7879053
- DOI: 10.1021/acs.jpcb.0c10489
Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations
Abstract
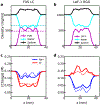
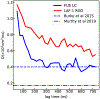
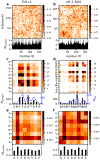
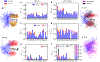
The formation of membraneless organelles in cells commonly occurs via liquid-liquid phase separation (LLPS) and is in many cases driven by multivalent interactions between intrinsically disordered proteins (IDPs). Investigating the nature of these interactions, and their effect on dynamics within the condensed phase, is therefore of critical importance but very challenging for either simulation or experiment. Here, we study these interactions and their dynamics by pairing a novel multiscale simulation strategy with microsecond all-atom MD simulations of a condensed, IDP-rich phase. We simulate two IDPs this way, the low complexity domain of FUS and the N-terminal disordered domain of LAF-1, and find good agreement with experimental information about average density, water content, and residue-residue contacts. We go significantly beyond what is known from experiments by showing that ion partitioning within the condensed phase is largely driven by the charge distribution of the proteins and-in the cases considered-shows little evidence of preferential interactions of the ions with the proteins. Furthermore, we can probe the microscopic diffusive dynamics within the condensed phase, showing that water and ions are in dynamic equilibrium between dense and dilute phases, and their diffusion is reduced in the dense phase. Despite their high concentration in the condensate, the protein molecules also remain mobile, explaining the observed liquid-like properties of this phase. We finally show that IDP self-association is driven by a combination of nonspecific hydrophobic interactions as well as hydrogen bonds, salt bridges, and π-π and cation-π interactions. The simulation approach presented here allows the structural and dynamical properties of biomolecular condensates to be studied in microscopic detail and is generally applicable to single- and multicomponent systems of proteins and nucleic acids involved in LLPS.
Figures

Similar articles
-
Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins.Proc Natl Acad Sci U S A. 2020 Nov 17;117(46):28795-28805. doi: 10.1073/pnas.2008122117. Epub 2020 Nov 2. Proc Natl Acad Sci U S A. 2020. PMID: 33139563 Free PMC article.
-
Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates.Biochemistry. 2018 May 1;57(17):2499-2508. doi: 10.1021/acs.biochem.8b00058. Epub 2018 Mar 13. Biochemistry. 2018. PMID: 29509422 Review.
-
Biological soft matter: intrinsically disordered proteins in liquid-liquid phase separation and biomolecular condensates.Essays Biochem. 2022 Dec 16;66(7):831-847. doi: 10.1042/EBC20220052. Essays Biochem. 2022. PMID: 36350034 Review.
-
Accurate model of liquid-liquid phase behavior of intrinsically disordered proteins from optimization of single-chain properties.Proc Natl Acad Sci U S A. 2021 Nov 2;118(44):e2111696118. doi: 10.1073/pnas.2111696118. Proc Natl Acad Sci U S A. 2021. PMID: 34716273 Free PMC article.
-
Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior.Proc Natl Acad Sci U S A. 2020 May 26;117(21):11421-11431. doi: 10.1073/pnas.2000223117. Epub 2020 May 11. Proc Natl Acad Sci U S A. 2020. PMID: 32393642 Free PMC article.
Cited by
-
Mechanistic insights into condensate formation of human liver-type phosphofructokinase by stochastic modeling approaches.Sci Rep. 2024 Aug 16;14(1):19011. doi: 10.1038/s41598-024-69534-w. Sci Rep. 2024. PMID: 39152221 Free PMC article.
-
Time-Dependent Material Properties of Aging Biomolecular Condensates from Different Viscoelasticity Measurements in Molecular Dynamics Simulations.J Phys Chem B. 2023 May 25;127(20):4441-4459. doi: 10.1021/acs.jpcb.3c01292. Epub 2023 May 17. J Phys Chem B. 2023. PMID: 37194953 Free PMC article.
-
Chemical tools for study and modulation of biomolecular phase transitions.Chem Sci. 2022 Nov 21;13(48):14226-14245. doi: 10.1039/d2sc04907d. eCollection 2022 Dec 14. Chem Sci. 2022. PMID: 36545140 Free PMC article. Review.
-
Aromatic and arginine content drives multiphasic condensation of protein-RNA mixtures.Biophys J. 2024 Jun 4;123(11):1342-1355. doi: 10.1016/j.bpj.2023.06.024. Epub 2023 Jul 5. Biophys J. 2024. PMID: 37408305 Free PMC article.
-
Toward Accurate Simulation of Coupling between Protein Secondary Structure and Phase Separation.J Am Chem Soc. 2024 Jan 10;146(1):342-357. doi: 10.1021/jacs.3c09195. Epub 2023 Dec 19. J Am Chem Soc. 2024. PMID: 38112495 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

