Multicenter Evaluation of the Clinical Performance and the Neutralizing Antibody Activity Prediction Properties of 10 High-Throughput Serological Assays Used in Clinical Laboratories
- PMID: 33303562
- PMCID: PMC8106733
- DOI: 10.1128/JCM.02511-20
Multicenter Evaluation of the Clinical Performance and the Neutralizing Antibody Activity Prediction Properties of 10 High-Throughput Serological Assays Used in Clinical Laboratories
Abstract
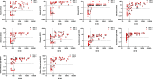
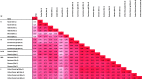
As the coronavirus disease 2019 (COVID-19) pandemic second wave is emerging, it is of the upmost importance to screen the population immunity in order to keep track of infected individuals. Consequently, immunoassays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with high specificity and positive predictive values are needed to obtain an accurate epidemiological picture. As more data accumulate about the immune responses and the kinetics of neutralizing-antibody (nAb) production in SARS-CoV-2-infected individuals, new applications are forecast for serological assays such as nAb activity prediction in convalescent-phase plasma from recovered patients. This multicenter study, involving six hospital centers, determined the baseline clinical performances, reproducibility, and nAb level correlations of 10 commercially available immunoassays. In addition, three lateral-flow chromatography assays were evaluated, as these devices can be used in logistically challenged areas. All assays were evaluated using the same patient panels in duplicate, thus enabling accurate comparison of the tests. Seven immunoassays examined in this study were shown to have excellent specificity (98 to 100%) and good to excellent positive predictive values (82 to 100%) when used in a low (5%)-seroprevalence setting. We observed sensitivities as low as 74% and as high as 95% at ≥15 days after symptom onset. The determination of optimized cutoff values through receiver operating characteristic (ROC) curve analyses had a significant impact on the diagnostic resolution of several enzyme immunoassays by increasing the sensitivity significantly without a large trade-off in specificity. We found that spike-based immunoassays seem to be better correlates of nAb activity. Finally, the results reported here will add to the general knowledge of the interlaboratory reproducibility of clinical performance parameters of immunoassays and provide new evidence about nAb activity prediction.
Keywords: COVID-19; SARS-CoV-2; antigen; immunoassays; neutralizing antibodies; serology.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi:10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Sironi M, Hasnain SE, Rosenthal B, Phan T, Luciani F, Shaw M-A, Sallum MA, Mirhashemi ME, Morand S, González-Candelas F, Editors of Infection, Genetics and Evolution. 2020. SARS-CoV-2 and COVID-19: a genetic, epidemiological, and evolutionary perspective. Infect Genet Evol 84:104384. doi:10.1016/j.meegid.2020.104384. - DOI - PMC - PubMed
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi:10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

