A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa
- PMID: 33304123
- PMCID: PMC7723883
- DOI: 10.3897/mycokeys.75.59872
A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa
Abstract
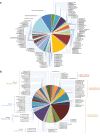
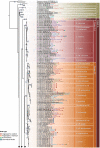
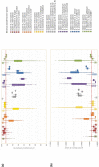
Ganoderma P. Karst. is a cosmopolitan genus of white-rot fungi which comprises species with highly-prized pharmaceutical properties, valuable biotechnological applications and of significant phytopathological interest. However, the status of the taxonomy within the genus is still highly controversial and ambiguous despite the progress made through molecular approaches. A metadata analysis of 3908 nuclear ribosomal internal transcribed spacer (ITS) rDNA sequences obtained from GenBank/ENA/DDBJ and UNITE was performed by targeting sequences annotated as Ganoderma, but also sequences from environmental samples and from material examined for the first time. Ganoderma taxa segregated into five main lineages (Clades A to E). Clade A corresponds to the core of laccate species and includes G. shanxiense and three major well-supported clusters: Cluster A.1 ('G. lucidum sensu lato') consists of taxa from Eurasia and North America, Cluster A.2 of material with worldwide occurrence including G. resinaceum and Cluster A.3 is composed of species originating from all continents except Europe and comprises G. lingzhi. Clade B includes G. applanatum and allied species with a Holarctic distribution. Clade C comprises taxa from Asia and Africa only. Clade D consists of laccate taxa with tropical/subtropical occurrence, while clade E harbours the highest number of non-laccate species with a cosmopolitan distribution. The 92 Ganoderma-associated names, initially used for sequences labelling, correspond to at least 80 taxa. Amongst them, 21 constitute putatively new phylospecies after our application of criteria relevant to the robustness/support of the terminal clades, intra- and interspecific genetic divergence and available biogeographic data. Moreover, several other groups or individual sequences seem to represent distinct taxonomic entities and merit further investigation. A particularly large number of the public sequences was revealed to be insufficiently and/or incorrectly identified, for example, 87% and 78% of entries labelled as G. australe and G. lucidum, respectively. In general, ITS demonstrated high efficacy in resolving relationships amongst most of the Ganoderma taxa; however, it was not equally useful at elucidating species barriers across the entire genus and such cases are outlined. Furthermore, we draw conclusions on biogeography by evaluating species occurrence on a global scale in conjunction with phylogenetic structure/patterns. The sequence variability assessed in ITS spacers could be further exploited for diagnostic purposes.
Keywords: Biogeography; ITS; fungal diversity; medicinal mushroom; phylogeny; taxonomy.
Vassiliki Fryssouli, Georgios I. Zervakis, Elias Polemis, Milton A. Typas.
Figures






References
-
- Adaskaveg JE, Gilbertson RL. (1986) Cultural studies and genetics of sexuality of Ganoderma lucidum and G. tsugae in relation to the taxonomy of the G. lucidum complex. Mycologia 78(5): 694–705. 10.1080/00275514.1986.12025312 - DOI
-
- Adaskaveg JE, Gilbertson RL. (1988) Basidiospores, pilocystidia, and other basidiocarp characters in several species of the G. lucidum complex. Mycologia 80(4): 493–507. 10.1080/00275514.1988.12025571 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
