Reverse Genetics System for Rabbit vesivirus
- PMID: 33304341
- PMCID: PMC7693663
- DOI: 10.3389/fmicb.2020.596245
Reverse Genetics System for Rabbit vesivirus
Abstract
Most caliciviruses are refractory to replication in cell culture and only a few members of the family propagate in vitro. Rabbit vesivirus (RaV) is unique due to its ability to grow to high titers in several animal and human cell lines. This outstanding feature makes RaV an ideal candidate for reverse genetics studies, an invaluable tool to understand the molecular basis of virus replication, the biological functions of viral genes and their roles in pathogenesis. The recovery of viruses from a cDNA clone is a prerequisite for reverse genetics studies. In this work, we constructed a RaV infectious cDNA clone using a plasmid expression vector, under the control of bacteriophage T7 RNA-polymerase promoter. The transfection of permissive cells with this plasmid DNA in the presence of T7 RNA-polymerase, provided in trans by a helper recombinant poxvirus, led to de novo synthesis of RNA transcripts that emulated the viral genome. The RaV progeny virus produced the typical virus-induced cytopathic effect after several passages of cell culture supernatants. Similarly, infectious RaV was recovered when the transcription step was performed in vitro, prior to transfection, provided that a 5'-cap structure was added to the 5' end of synthetic genome-length RNAs. In this work, we report an efficient and consistent RaV rescue system based on a cDNA transcription vector, as a tool to investigate calicivirus biology through reverse genetics.
Keywords: Vesivirus; calicivirus; infectious cDNA clone; reverse genetics; virus rescue.
Copyright © 2020 Álvarez, García-Manso, Dalton, Martín-Alonso, Nicieza, Podadera, Acosta-Zaldívar, de Llano and Parra.
Figures

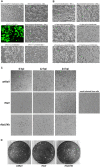



References
-
- Bridgen A., Elliot R. M. (2000). “Chapter 9. reverse genetics of RNA viruses,” in RNA Viruses: A Practical Approach, 1st Edn, ed. Cann A. J. (New York, NY: Oxford University Press; ), 201–227.
LinkOut - more resources
Full Text Sources
Miscellaneous

