M1 Polarization Markers Are Upregulated in Basal-Like Breast Cancer Molecular Subtype and Associated With Favorable Patient Outcome
- PMID: 33304345
- PMCID: PMC7701279
- DOI: 10.3389/fimmu.2020.560074
M1 Polarization Markers Are Upregulated in Basal-Like Breast Cancer Molecular Subtype and Associated With Favorable Patient Outcome
Abstract
Background: Breast cancer heterogeneity is an essential element that plays a role in the therapy response variability and the patient's outcome. This highlights the need for more precise subtyping methods that focus not only on tumor cells but also investigate the profile of stromal cells as well as immune cells.
Objectives: To mine publicly available transcriptomic breast cancer datasets and reanalyze their transcriptomic profiling using unsupervised clustering in order to identify novel subsets in molecular subtypes of breast cancer, then explore the stromal and immune cells profile in each subset using bioinformatics and systems immunology approaches.
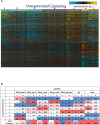
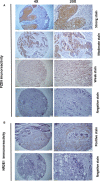
Materials and methods: Transcriptomic data from 1,084 breast cancer patients obtained from The Cancer Genome Atlas (TCGA) database were extracted and subjected to unsupervised clustering using a recently described, multi-step algorithm called Iterative Clustering and Guide-gene Selection (ICGS). For each cluster, the stromal and immune profile was investigated using ESTIMATE and CIBERSORT analytical tool. Clinical outcomes and differentially expressed genes of the characterized clusters were identified and validated in silico and in vitro in a cohort of 80 breast cancer samples by immunohistochemistry.
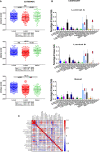
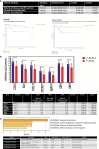
Results: Seven unique sub-clusters showed distinct molecular and clinical profiles between the well-known breast cancer subtypes. Those unsupervised clusters identified more homogenous subgroups in each of the classical subtypes with a different prognostic profile. Immune profiling of the identified clusters showed that while the classically activated macrophages (M1) are correlated with the more aggressive basal-like breast cancer subtype, the alternatively activated macrophages (M2) showed a higher level of infiltration in luminal A and luminal B subtypes. Indeed, patients with higher levels of M1 expression showed less advanced disease and better patient outcomes presented as prolonged overall survival. Moreover, the M1 high basal-like breast cancer group showed a higher expression of interferon-gamma induced chemokines and guanylate-binding proteins (GBPs) involved in immunity against microbes.
Conclusion: Adding immune profiling using transcriptomic data can add precision for diagnosis and prognosis and can cluster patients according to the available modalities of therapy in a more personalized approach.
Keywords: basal like; breast cancer; macrophages; transcriptomic; tumor infiltrated immune cells.
Copyright © 2020 Hachim, Hachim, Talaat, Yakout and Hamoudi.
Figures



References
-
- Peart O. Metastatic Breast Cancer. Radiologic Technol (2017) 88(5):519m–39m. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

