Homology modeling in the time of collective and artificial intelligence
- PMID: 33304450
- PMCID: PMC7695898
- DOI: 10.1016/j.csbj.2020.11.007
Homology modeling in the time of collective and artificial intelligence
Abstract
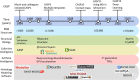
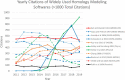
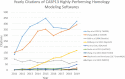
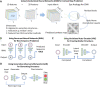
Homology modeling is a method for building protein 3D structures using protein primary sequence and utilizing prior knowledge gained from structural similarities with other proteins. The homology modeling process is done in sequential steps where sequence/structure alignment is optimized, then a backbone is built and later, side-chains are added. Once the low-homology loops are modeled, the whole 3D structure is optimized and validated. In the past three decades, a few collective and collaborative initiatives allowed for continuous progress in both homology and ab initio modeling. Critical Assessment of protein Structure Prediction (CASP) is a worldwide community experiment that has historically recorded the progress in this field. Folding@Home and Rosetta@Home are examples of crowd-sourcing initiatives where the community is sharing computational resources, whereas RosettaCommons is an example of an initiative where a community is sharing a codebase for the development of computational algorithms. Foldit is another initiative where participants compete with each other in a protein folding video game to predict 3D structure. In the past few years, contact maps deep machine learning was introduced to the 3D structure prediction process, adding more information and increasing the accuracy of models significantly. In this review, we will take the reader in a journey of exploration from the beginnings to the most recent turnabouts, which have revolutionized the field of homology modeling. Moreover, we discuss the new trends emerging in this rapidly growing field.
Keywords: Artificial intelligence; Collective intelligence; Homology modeling; Machine learning; Protein 3D structure; Structural bioinformatics.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article.
-
Critical Assessment of Methods for Predicting the 3D Structure of Proteins and Protein Complexes.Annu Rev Biophys. 2023 May 9;52:183-206. doi: 10.1146/annurev-biophys-102622-084607. Epub 2023 Jan 10. Annu Rev Biophys. 2023. PMID: 36626764 Free PMC article. Review.
-
Protein structure modeling for CASP10 by multiple layers of global optimization.Proteins. 2014 Feb;82 Suppl 2:188-95. doi: 10.1002/prot.24397. Epub 2013 Oct 24. Proteins. 2014. PMID: 23966235
-
Modeling structurally variable regions in homologous proteins with rosetta.Proteins. 2004 May 15;55(3):656-77. doi: 10.1002/prot.10629. Proteins. 2004. PMID: 15103629
-
A glance into the evolution of template-free protein structure prediction methodologies.Biochimie. 2020 Aug;175:85-92. doi: 10.1016/j.biochi.2020.04.026. Epub 2020 May 15. Biochimie. 2020. PMID: 32417458 Review.
Cited by
-
SARS-CoV-2 proteins structural studies using synchrotron radiation.Biophys Rev. 2023 Sep 29;15(5):1185-1194. doi: 10.1007/s12551-023-01153-7. eCollection 2023 Oct. Biophys Rev. 2023. PMID: 37974992 Free PMC article. Review.
-
The Application of MD Simulation to Lead Identification, Vaccine Design, and Structural Studies in Combat against Leishmaniasis - A Review.Mini Rev Med Chem. 2024;24(11):1089-1111. doi: 10.2174/1389557523666230901105231. Mini Rev Med Chem. 2024. PMID: 37680156 Review.
-
A Comparative Analysis of Cockroach and Mosquito, Octopamine Receptor Homologues Produced Using Chimera, Swiss-Model, and AlphaFold Molecular Modeling Tools.ACS Omega. 2025 Feb 19;10(8):7907-7919. doi: 10.1021/acsomega.4c08755. eCollection 2025 Mar 4. ACS Omega. 2025. PMID: 40060804 Free PMC article.
-
Comparing models and experimental structures of the GPR101 receptor: Artificial intelligence yields highly accurate models.J Mol Graph Model. 2025 Nov;140:109103. doi: 10.1016/j.jmgm.2025.109103. Epub 2025 Jun 3. J Mol Graph Model. 2025. PMID: 40472416
-
Characterization of a Stable Form of Carboxypeptidase G2 (Glucarpidase), a Potential Biobetter Variant, From Acinetobacter sp. 263903-1.Mol Biotechnol. 2021 Dec;63(12):1155-1168. doi: 10.1007/s12033-021-00370-3. Epub 2021 Jul 15. Mol Biotechnol. 2021. PMID: 34268672
References
-
- Hargittai I. Linus Pauling’s quest for the structure of proteins. Struct. Chem. 2009;21(1):1–7.
-
- Muhammed M.T., Aki-Yalcin E. Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019;93(1):12–20. - PubMed
-
- Hatfield M.P., Lovas S. Conformational sampling techniques. Curr. Pharm. Des. 2014;20(20):3303–3313. - PubMed
-
- Moult J. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23(3):2–4. - PubMed
-
- Samuel A.L. Some Studies in Machine Learning Using the Game of Checkers. IBM J. Res. Dev. 1959;3(3):210–229.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
