gauseR: Simple methods for fitting Lotka-Volterra models describing Gause's "Struggle for Existence"
- PMID: 33304536
- PMCID: PMC7713957
- DOI: 10.1002/ece3.6926
gauseR: Simple methods for fitting Lotka-Volterra models describing Gause's "Struggle for Existence"
Abstract
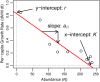
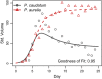
Point 1: The ecological models of Alfred J. Lotka and Vito Volterra have had an enormous impact on ecology over the past century. Some of the earliest-and clearest-experimental tests of these models were famously conducted by Georgy Gause in the 1930s. Although well known, the data from these experiments are not widely available and are often difficult to analyze using standard statistical and computational tools. Point 2: Here, we introduce the gauseR package, a collection of tools for fitting Lotka-Volterra models to time series data of one or more species. The package includes several methods for parameter estimation and optimization, and includes 42 datasets from Gause's species interaction experiments and related work. Additionally, we include with this paper a short blog post discussing the historical importance of these data and models, and an R vignette with a walk-through introducing the package methods. The package is available for download at github.com/adamtclark/gauseR. Point 3: To demonstrate the package, we apply it to several classic experimental studies from Gause, as well as two other well-known datasets on multi-trophic dynamics on Isle Royale, and in spatially structured mite populations. In almost all cases, models fit observations closely and fitted parameter values make ecological sense. Point 4: Taken together, we hope that the methods, data, and analyses that we present here provide a simple and user-friendly way to interact with complex ecological data. We are optimistic that these methods will be especially useful to students and educators who are studying ecological dynamics, as well as researchers who would like a fast tool for basic analyses.
Keywords: competition; differential equation; growth rate; logistic growth; optimization; predator‐prey.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts related to the research, code, or data presented in this paper.
Figures




References
-
- Carrara, F. , Giometto, A. , Seymour, M. , Rinaldo, A. , & Altermatt, F. (2015). Inferring Species Interactions in Ecological Communities: A Comparison of Methods at Different Levels of Complexity. Methods in Ecology and Evolution, 6, 895–906. 10.1111/2041-210X.12363 - DOI
-
- Case, T. J. , & Bender, E. A. (1981). Testing for Higher Order Interactions. The American Naturalist, 118, 920–929. 10.1086/283885 - DOI
-
- Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics, 31, 343–366. 10.1146/annurev.ecolsys.31.1.343 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

