Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview
- PMID: 33304888
- PMCID: PMC7693651
- DOI: 10.3389/fbioe.2020.570828
Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview
Abstract
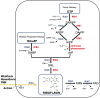
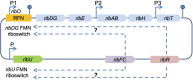
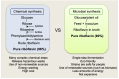
Riboflavin is a crucial micronutrient that is a precursor to coenzymes flavin mononucleotide and flavin adenine dinucleotide, and it is required for biochemical reactions in all living cells. For decades, one of the most important applications of riboflavin has been its global use as an animal and human nutritional supplement. Being well-informed of the latest research on riboflavin production via the fermentation process is necessary for the development of new and improved microbial strains using biotechnology and metabolic engineering techniques to increase vitamin B2 yield. In this review, we describe well-known industrial microbial producers, namely, Ashbya gossypii, Bacillus subtilis, and Candida spp. and summarize their biosynthetic pathway optimizations through genetic and metabolic engineering, combined with random chemical mutagenesis and rational medium components to increase riboflavin production.
Keywords: food/feed additive; genetically modified microorganisms; metabolic engineering; riboflavin; vitamin B2.
Copyright © 2020 Averianova, Balabanova, Son, Podvolotskaya and Tekutyeva.
Figures
References
-
- Abd-Alla M. H., Bagy M. M. K., Nafady N. A., Morsy F. M., Mahmoud A. E. (2016). Activation of riboflavin production by Bacillus subtilis (KU559874) and Bacillus tequilensis (KU559876). EC Bacteriol. Virol. Res. 2 131–150.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

