Selective Chemical Functionalization at N6-Methyladenosine Residues in DNA Enabled by Visible-Light-Mediated Photoredox Catalysis
- PMID: 33305571
- PMCID: PMC7760100
- DOI: 10.1021/jacs.0c10616
Selective Chemical Functionalization at N6-Methyladenosine Residues in DNA Enabled by Visible-Light-Mediated Photoredox Catalysis
Abstract
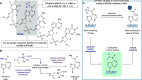
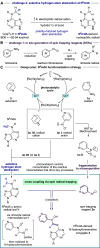
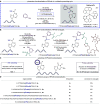
Selective chemistry that modifies the structure of DNA and RNA is essential to understanding the role of epigenetic modifications. We report a visible-light-activated photocatalytic process that introduces a covalent modification at a C(sp3)-H bond in the methyl group of N6-methyl deoxyadenosine and N6-methyl adenosine, epigenetic modifications of emerging importance. A carefully orchestrated reaction combines reduction of a nitropyridine to form a nitrosopyridine spin-trapping reagent and an exquisitely selective tertiary amine-mediated hydrogen-atom abstraction at the N6-methyl group to form an α-amino radical. Cross-coupling of the putative α-amino radical with nitrosopyridine leads to a stable conjugate, installing a label at N6-methyl-adenosine. We show that N6-methyl deoxyadenosine-containing oligonucleotides can be enriched from complex mixtures, paving the way for applications to identify this modification in genomic DNA and RNA.
Conflict of interest statement
The authors declare the following competing financial interest(s): S.B. is an advisor and shareholder of Cambridge Epigenetix Ltd. A patent application is pending.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

