P53 induces senescence in the unstable progeny of aneuploid cells
- PMID: 33305692
- PMCID: PMC7781663
- DOI: 10.1080/15384101.2020.1850968
P53 induces senescence in the unstable progeny of aneuploid cells
Abstract
Aneuploidy is the condition of having an imbalanced karyotype, which is associated with tumor initiation, evolution, and acquisition of drug-resistant features, possibly by generating heterogeneous populations of cells with distinct genotypes and phenotypes. Multicellular eukaryotes have therefore evolved a range of extrinsic and cell-autonomous mechanisms for restraining proliferation of aneuploid cells, including activation of the tumor suppressor protein p53. However, accumulating evidence indicates that a subset of aneuploid cells can escape p53-mediated growth restriction and continue proliferating in vitro. Here we show that such aneuploid cell lines display a robust modal karyotype and low frequency of chromosomal aberrations despite ongoing chromosome instability. Indeed, while these aneuploid cells are able to survive for extended periods in vitro, their chromosomally unstable progeny remain subject to p53-induced senescence and growth restriction, leading to subsequent elimination from the aneuploid pool. This mechanism helps maintain low levels of heterogeneity in aneuploid populations and may prevent detrimental evolutionary processes such as cancer progression and development of drug resistance.
Keywords: P53; aneuploidy; genome instability; senescence.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

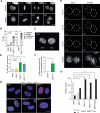
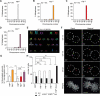
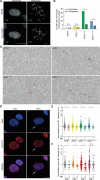
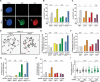
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
