Development and Validation of a Tool Integrating the 21-Gene Recurrence Score and Clinical-Pathological Features to Individualize Prognosis and Prediction of Chemotherapy Benefit in Early Breast Cancer
- PMID: 33306425
- PMCID: PMC8078482
- DOI: 10.1200/JCO.20.03007
Development and Validation of a Tool Integrating the 21-Gene Recurrence Score and Clinical-Pathological Features to Individualize Prognosis and Prediction of Chemotherapy Benefit in Early Breast Cancer
Abstract
Purpose: The 21-gene recurrence score (RS) is prognostic for distant recurrence (DR) and predictive for chemotherapy benefit in early breast cancer, whereas clinical-pathological factors are only prognostic. Integration of genomic and clinical features offers the potential to guide adjuvant chemotherapy use with greater precision.
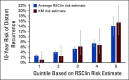
Methods: We developed a new tool (RSClin) that integrates RS with tumor grade, tumor size, and age using a patient-specific meta-analysis including 10,004 women with hormone receptor-positive, human epidermal growth factor receptor 2-negative, and node-negative breast cancer who received endocrine therapy alone in the B-14 (n = 577) and TAILORx (n = 4,854) trials or plus chemotherapy in TAILORx (n = 4,573). Cox models for RSClin were compared with RS alone and clinical-pathological features alone using likelihood ratio tests. RSClin estimates of DR used a baseline risk with TAILORx event rates to reflect current medical practice. A patient-specific estimator of absolute chemotherapy benefit was computed using individualized relative chemotherapy effect from the randomized TAILORx and B-20 trials. External validation of risk estimation was performed by comparing RSClin estimated risk and observed risk in 1,098 women in the Clalit registry.
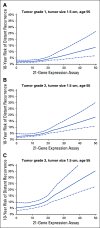
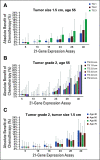
Results: RSClin provides more prognostic information (likelihood ratio χ2) for DR than RS or clinical-pathological factors alone (both P < .001, likelihood ratio test). In external validation, the RSClin risk estimate was prognostic for DR risk in the Clalit registry (P < .001) and the estimated risk closely approximated the observed 10-year risk (Lin concordance 0.962). The absolute chemotherapy benefit estimate ranges from 0% to 15% as the RS ranges from 11 to 50 using RSClin in a 55-year-old woman with a 1.5-cm intermediate-grade tumor.
Conclusion: The RSClin tool integrates clinical-pathological and genomic risk to guide adjuvant chemotherapy in node-negative breast cancer and provides more individualized information than clinical-pathological or genomic data alone.
Figures

Comment in
-
Better Together: Clinical and Genomic Data to Inform Shared Decision Making.J Clin Oncol. 2021 Feb 20;39(6):545-547. doi: 10.1200/JCO.20.03234. Epub 2020 Dec 11. J Clin Oncol. 2021. PMID: 33306424 No abstract available.
-
What Is the Optimal Model to Estimate the Benefits of Chemotherapy in Patients With Hormone Receptor-Positive, HER2-Negative, Node-Negative Breast Cancer?J Clin Oncol. 2021 Jun 10;39(17):1946-1947. doi: 10.1200/JCO.21.00178. Epub 2021 Apr 1. J Clin Oncol. 2021. PMID: 33793293 No abstract available.
-
Reply to K. Ando et al.J Clin Oncol. 2021 Jun 10;39(17):1947-1948. doi: 10.1200/JCO.21.00424. Epub 2021 Apr 1. J Clin Oncol. 2021. PMID: 33793318 Free PMC article. No abstract available.
References
-
- Paik S Shak S Tang G, et al. : A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817-2826, 2004 - PubMed
-
- Paik S Tang G Shak S, et al. : Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24:3726-3734, 2006 - PubMed
-
- Albain KS Barlow WE Shak S, et al. : Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol 11:55-65, 2010 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

