Principles of Self-Organization of the Mammalian Embryo
- PMID: 33306953
- PMCID: PMC8212876
- DOI: 10.1016/j.cell.2020.11.003
Principles of Self-Organization of the Mammalian Embryo
Abstract
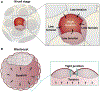
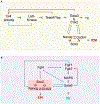
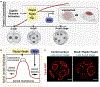
Early embryogenesis is a conserved and self-organized process. In the mammalian embryo, the potential for self-organization is manifested in its extraordinary developmental plasticity, allowing a correctly patterned embryo to arise despite experimental perturbation. The underlying mechanisms enabling such regulative development have long been a topic of study. In this Review, we summarize our current understanding of the self-organizing principles behind the regulative nature of the early mammalian embryo. We argue that geometrical constraints, feedback between mechanical and biochemical factors, and cellular heterogeneity are all required to ensure the developmental plasticity of mammalian embryo development.
Copyright © 2020. Published by Elsevier Inc.
Figures


References
-
- Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, and Yamanaka Y (2014). Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 141, 2813–2824. - PubMed
-
- Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, and Chazaud C (2014). Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development 141, 3637–3648. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

