D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization
- PMID: 33306985
- PMCID: PMC7707640
- DOI: 10.1016/j.chom.2020.11.012
D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization
Abstract
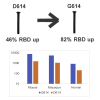
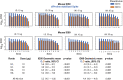
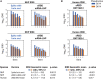
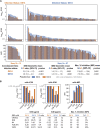
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein acquired a D614G mutation early in the pandemic that confers greater infectivity and is now the globally dominant form. To determine whether D614G might also mediate neutralization escape that could compromise vaccine efficacy, sera from spike-immunized mice, nonhuman primates, and humans were evaluated for neutralization of pseudoviruses bearing either D614 or G614 spike. In all cases, the G614 pseudovirus was moderately more susceptible to neutralization. The G614 pseudovirus also was more susceptible to neutralization by receptor-binding domain (RBD) monoclonal antibodies and convalescent sera from people infected with either form of the virus. Negative stain electron microscopy revealed a higher percentage of the 1-RBD "up" conformation in the G614 spike, suggesting increased epitope exposure as a mechanism of enhanced vulnerability to neutralization. Based on these findings, the D614G mutation is not expected to be an obstacle for current vaccine development.
Keywords: COVID-19; D614G; LNP; SARS-CoV-2; Spike; electron micrograph; mRNA; neutralization; nucleoside-modified; vaccine.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that N.P. and D.W. are named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins and vaccines. We have disclosed those interests fully to the University of Pennsylvania, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents.
Figures

References
-
- Alameh M.G., Weissman D., Pardi N. Messenger RNA-Based Vaccines Against Infectious Diseases. Curr. Top. Microbiol. Immunol. 2020 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

