Transcriptomic Profiling of Human Pluripotent Stem Cell-derived Retinal Pigment Epithelium over Time
- PMID: 33307245
- PMCID: PMC8602392
- DOI: 10.1016/j.gpb.2020.08.002
Transcriptomic Profiling of Human Pluripotent Stem Cell-derived Retinal Pigment Epithelium over Time
Abstract
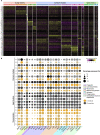
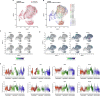
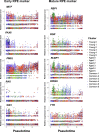
Human pluripotent stem cell (hPSC)-derived progenies are immature versions of cells, presenting a potential limitation to the accurate modelling of diseases associated with maturity or age. Hence, it is important to characterise how closely cells used in culture resemble their native counterparts. In order to select appropriate time points of retinal pigment epithelium (RPE) cultures that reflect native counterparts, we characterised the transcriptomic profiles of the hPSC-derived RPE cells from 1- and 12-month cultures. We differentiated the human embryonic stem cell line H9 into RPE cells, performed single-cell RNA-sequencing of a total of 16,576 cells to assess the molecular changes of the RPE cells across these two culture time points. Our results indicate the stability of the RPE transcriptomic signature, with no evidence of an epithelial-mesenchymal transition, and with the maturing populations of the RPE observed with time in culture. Assessment of Gene Ontology pathways revealed that as the cultures age, RPE cells upregulate expression of genes involved in metal binding and antioxidant functions. This might reflect an increased ability to handle oxidative stress as cells mature. Comparison with native human RPE data confirms a maturing transcriptional profile of RPE cells in culture. These results suggest that long-term in vitro culture of RPE cells allows the modelling of specific phenotypes observed in native mature tissues. Our work highlights the transcriptional landscape of hPSC-derived RPE cells as they age in culture, which provides a reference for native and patient samples to be benchmarked against.
Keywords: Ageing; Human embryonic stem cell; Human pluripotent stem cell; Retinal pigment epithelium; Single-cell RNA sequencing.
Copyright © 2021 Beijing Institute of Genomics. Published by Elsevier B.V. All rights reserved.
Figures









References
-
- Perez V.L., Saeed A.M., Tan Y., Urbieta M., Cruz-Guilloty F. The eye: a window to the soul of the immune system. J Autoimmun. 2013;45:7–14. - PubMed
-
- Xu H., Chen M., Forrester J.V. Para-inflammation in the aging retina. Prog Retin Eye Res. 2009;28:348–368. - PubMed
-
- Lidgerwood G.E., Morris A.J., Conquest A., Daniszewski M., Rooney L.A., Lim S.Y., et al. Role of lysophosphatidic acid in the retinal pigment epithelium and photoreceptors. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:750–761. - PubMed
-
- Lidgerwood G.E., Lim S.Y., Crombie D.E., Ali R., Gill K.P., Hernández D., et al. Defined medium conditions for the induction and expansion of human pluripotent stem cell-derived retinal pigment epithelium. Stem Cell Rev Rep. 2016;12:179–188. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

