Identification of CD8+ T cell epitopes through proteasome cleavage site predictions
- PMID: 33308150
- PMCID: PMC7733697
- DOI: 10.1186/s12859-020-03782-1
Identification of CD8+ T cell epitopes through proteasome cleavage site predictions
Abstract
Background: We previously introduced PCPS (Proteasome Cleavage Prediction Server), a web-based tool to predict proteasome cleavage sites using n-grams. Here, we evaluated the ability of PCPS immunoproteasome cleavage model to discriminate CD8+ T cell epitopes.
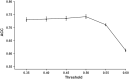
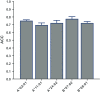
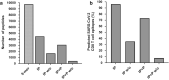
Results: We first assembled an epitope dataset consisting of 844 unique virus-specific CD8+ T cell epitopes and their source proteins. We then analyzed cleavage predictions by PCPS immunoproteasome cleavage model on this dataset and compared them with those provided by a related method implemented by NetChop web server. PCPS was clearly superior to NetChop in term of sensitivity (0.89 vs. 0.79) but somewhat inferior with regard to specificity (0.55 vs. 0.60). Judging by the Mathew's Correlation Coefficient, PCPS predictions were overall superior to those provided by NetChop (0.46 vs. 0.39). We next analyzed the power of C-terminal cleavage predictions provided by the same PCPS model to discriminate CD8+ T cell epitopes, finding that they could be discriminated from random peptides with an accuracy of 0.74. Following these results, we tuned the PCPS web server to predict CD8+ T cell epitopes and predicted the entire SARS-CoV-2 epitope space.
Conclusions: We report an improved version of PCPS named iPCPS for predicting proteasome cleavage sites and peptides with CD8+ T cell epitope features. iPCPS is available for free public use at https://imed.med.ucm.es/Tools/pcps/ .
Keywords: CD8+ T cell epitope; Immunoproteasome; Peptide; Prediction; Proteasome; SARS-CoV-2.
Conflict of interest statement
All the authors declare that they have no competing interests.
Figures


References
-
- Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786(2):153–177. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

