A Molecular Calcium Integrator Reveals a Striatal Cell Type Driving Aversion
- PMID: 33308478
- PMCID: PMC9839359
- DOI: 10.1016/j.cell.2020.11.015
A Molecular Calcium Integrator Reveals a Striatal Cell Type Driving Aversion
Abstract
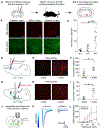
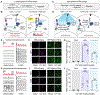
The ability to record transient cellular events in the DNA or RNA of cells would enable precise, large-scale analysis, selection, and reprogramming of heterogeneous cell populations. Here, we report a molecular technology for stable genetic tagging of cells that exhibit activity-related increases in intracellular calcium concentration (FLiCRE). We used FLiCRE to transcriptionally label activated neural ensembles in the nucleus accumbens of the mouse brain during brief stimulation of aversive inputs. Using single-cell RNA sequencing, we detected FLiCRE transcripts among the endogenous transcriptome, providing simultaneous readout of both cell-type and calcium activation history. We identified a cell type in the nucleus accumbens activated downstream of long-range excitatory projections. Taking advantage of FLiCRE's modular design, we expressed an optogenetic channel selectively in this cell type and showed that direct recruitment of this otherwise genetically inaccessible population elicits behavioral aversion. The specificity and minute resolution of FLiCRE enables molecularly informed characterization, manipulation, and reprogramming of activated cellular ensembles.
Keywords: aversion; calcium; light; neural activity; optogenetics; single-cell RNA sequencing; striatum; transcription factor.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A.Y.T. and M.I.S. have filed a patent covering some components used in this study (US provisional application 62/906,373; CZB file CZB-123S-P1; Stanford file S19-269; KT file 103182-1132922-002400PR).
Figures





Comment in
-
An expanding molecular toolbox untangles neural circuits.Nature. 2021 Nov;599(7884):335-337. doi: 10.1038/d41586-021-03054-9. Nature. 2021. PMID: 34754087 No abstract available.
References
-
- Anzalone AV, Koblan LW, and Liu DR (2020). Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nature biotechnology, 1–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

