Actin-Membrane Release Initiates Cell Protrusions
- PMID: 33308479
- PMCID: PMC7908823
- DOI: 10.1016/j.devcel.2020.11.024
Actin-Membrane Release Initiates Cell Protrusions
Abstract
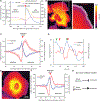
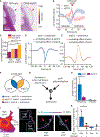
Despite the well-established role of actin polymerization as a driving mechanism for cell protrusion, upregulated actin polymerization alone does not initiate protrusions. Using a combination of theoretical modeling and quantitative live-cell imaging experiments, we show that local depletion of actin-membrane links is needed for protrusion initiation. Specifically, we show that the actin-membrane linker ezrin is depleted prior to protrusion onset and that perturbation of ezrin's affinity for actin modulates protrusion frequency and efficiency. We also show how actin-membrane release works in concert with actin polymerization, leading to a comprehensive model for actin-driven shape changes. Actin-membrane release plays a similar role in protrusions driven by intracellular pressure. Thus, our findings suggest that protrusion initiation might be governed by a universal regulatory mechanism, whereas the mechanism of force generation determines the shape and expansion properties of the protrusion.
Keywords: Brownian ratchet model; actin dynamics; cytoskeleton; intracellular force; lamellipodium; morphology; polymerization; protrusion; shape change.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Comment in
-
Letting Go to Move On: Membrane Detachment Initiates Protrusion.Dev Cell. 2020 Dec 21;55(6):671-672. doi: 10.1016/j.devcel.2020.11.026. Dev Cell. 2020. PMID: 33352139
References
-
- Andrew N, and Insall RH (2007). Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol 9, 193–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

