Using metacommunity ecology to understand environmental metabolomes
- PMID: 33311510
- PMCID: PMC7732844
- DOI: 10.1038/s41467-020-19989-y
Using metacommunity ecology to understand environmental metabolomes
Abstract
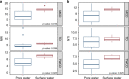
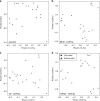
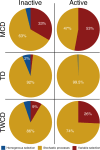
Environmental metabolomes are fundamentally coupled to microbially-linked biogeochemical processes within ecosystems. However, significant gaps exist in our understanding of their spatiotemporal organization, limiting our ability to uncover transferrable principles and predict ecosystem function. We propose that a theoretical paradigm, which integrates concepts from metacommunity ecology, is necessary to reveal underlying mechanisms governing metabolomes. We call this synthesis between ecology and metabolomics 'meta-metabolome ecology' and demonstrate its utility using a mass spectrometry dataset. We developed three relational metabolite dendrograms using molecular properties and putative biochemical transformations and performed ecological null modeling. Based upon null modeling results, we show that stochastic processes drove molecular properties while biochemical transformations were structured deterministically. We further suggest that potentially biochemically active metabolites were more deterministically assembled than less active metabolites. Understanding variation in the influences of stochasticity and determinism provides a way to focus attention on which meta-metabolomes and which parts of meta-metabolomes are most likely to be important to consider in mechanistic models. We propose that this paradigm will allow researchers to study the connections between ecological systems and their molecular processes in previously inaccessible detail.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Sengupta A, et al. Spatial gradients in the characteristics of soil-carbon fractions are associated with abiotic features but not microbial communities. Biogeosciences. 2019;16:3911–3928. doi: 10.5194/bg-16-3911-2019. - DOI
-
- Garayburu-Caruso, V. et al. Carbon limitation leads to thermodynamic regulation of aerobic metabolism. bioRxiv10.1101/2020.01.15.905331 (2020).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

