Development and validation of a three-long noncoding RNA signature for predicting prognosis of patients with gastric cancer
- PMID: 33311941
- PMCID: PMC7701940
- DOI: 10.3748/wjg.v26.i44.6929
Development and validation of a three-long noncoding RNA signature for predicting prognosis of patients with gastric cancer
Abstract
Background: Gastric cancer (GC) is one of the most frequently diagnosed gastrointestinal cancers throughout the world. Novel prognostic biomarkers are required to predict the prognosis of GC.
Aim: To identify a multi-long noncoding RNA (lncRNA) prognostic model for GC.
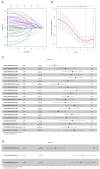
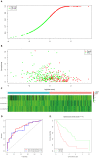
Methods: Transcriptome data and clinical data were downloaded from The Cancer Genome Atlas. COX and least absolute shrinkage and selection operator regression analyses were performed to screen for prognosis associated lncRNAs. Receiver operating characteristic curve and Kaplan-Meier survival analyses were applied to evaluate the effectiveness of the model.
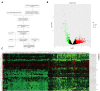
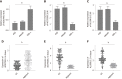
Results: The prediction model was established based on the expression of AC007991.4, AC079385.3, and AL109615.2 Based on the model, GC patients were divided into "high risk" and "low risk" groups to compare the differences in survival. The model was re-evaluated with the clinical data of our center.
Conclusion: The 3-lncRNA combination model is an independent prognostic factor for GC.
Keywords: Gastric cancer; Least absolute shrinkage and selection operator; Long noncoding RNA; Prognosis; Survival analysis.
©The Author(s) 2020. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: No potential conflicts of interest are disclosed.
Figures
Similar articles
-
Identification of a 6-lncRNA prognostic signature based on microarray re-annotation in gastric cancer.Cancer Med. 2020 Jan;9(1):335-349. doi: 10.1002/cam4.2621. Epub 2019 Nov 19. Cancer Med. 2020. PMID: 31743579 Free PMC article.
-
A prognostic 3-long noncoding RNA signature for patients with gastric cancer.J Cell Biochem. 2018 Nov;119(11):9261-9269. doi: 10.1002/jcb.27195. Epub 2018 Aug 3. J Cell Biochem. 2018. PMID: 30074647
-
Identification of a novel m6A-related lncRNA pair signature for predicting the prognosis of gastric cancer patients.BMC Gastroenterol. 2022 Feb 21;22(1):76. doi: 10.1186/s12876-022-02159-3. BMC Gastroenterol. 2022. PMID: 35189810 Free PMC article.
-
A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma.J Cell Biochem. 2019 Jan;120(1):213-223. doi: 10.1002/jcb.27321. Epub 2018 Sep 11. J Cell Biochem. 2019. PMID: 30206981
-
Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma.World J Gastroenterol. 2019 Jan 14;25(2):220-232. doi: 10.3748/wjg.v25.i2.220. World J Gastroenterol. 2019. PMID: 30670911 Free PMC article.
Cited by
-
N7-methylguanosine-related lncRNAs: Predicting the prognosis and diagnosis of colorectal cancer in the cold and hot tumors.Front Genet. 2022 Jul 22;13:952836. doi: 10.3389/fgene.2022.952836. eCollection 2022. Front Genet. 2022. PMID: 35937987 Free PMC article.
-
Development of an immunogenic cell death-related lncRNAs signature for prognostic risk assessment in gastric cancer.Transl Cancer Res. 2024 Aug 31;13(8):4420-4440. doi: 10.21037/tcr-24-344. Epub 2024 Aug 27. Transl Cancer Res. 2024. PMID: 39262480 Free PMC article.
-
Derivation, Comprehensive Analysis, and Assay Validation of a Pyroptosis-Related lncRNA Prognostic Signature in Patients With Ovarian Cancer.Front Oncol. 2022 Feb 24;12:780950. doi: 10.3389/fonc.2022.780950. eCollection 2022. Front Oncol. 2022. PMID: 35280739 Free PMC article.
-
Necroptosis-associated long noncoding RNAs can predict prognosis and differentiate between cold and hot tumors in ovarian cancer.Front Oncol. 2022 Jul 28;12:967207. doi: 10.3389/fonc.2022.967207. eCollection 2022. Front Oncol. 2022. PMID: 35965557 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. - PubMed
-
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. - PubMed
-
- Zhang T, Piao HY, Guo S, Zhao Y, Wang Y, Zheng ZC, Zhang J. LncRNA PCGEM1 enhances metastasis and gastric cancer invasion through targeting of miR-129-5p to regulate P4HA2 expression. Exp Mol Pathol. 2020;116:104487. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

