Whole-exome sequencing identifies prognostic mutational signatures in gastric cancer
- PMID: 33313229
- PMCID: PMC7729362
- DOI: 10.21037/atm-20-6620
Whole-exome sequencing identifies prognostic mutational signatures in gastric cancer
Abstract
Background: Gastric cancer (GC) is a heterogeneous disease, and is a leading cause of cancer deaths in Eastern Asia. Genomic analysis, such as whole-exome sequencing (WES), can help identify key genetic alterations leading to the malignancy and diversity of GC, and may help identify new drug targets.
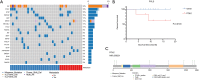
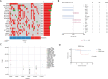
Methods: We identified genomic alterations in a cohort of 38 GC patients, including 26 metastatic and 12 non-metastatic patients. We analyzed the association between novel gene mutations and copy number variations (CNVs) with tumor metastasis and patient survival.
Results: A number of significantly mutated genes in somatic and germline cells were identified. Among them, ATAD3B somatic mutation, a potential biomarker of immunotherapy in stomach cancers, was associated with better patient survival (P=0.0939) and metastasis (P=0.074). POLE germline variation was correlated with shorter overall survival (OS; P=0.0100). Novel CNVs were also identified and can potentially be used as biomarkers. These included 9p24.1 deletion (P=0.0376) and 16p11.2 amplification (P=0.0066), which were both associated with shorter OS. CNVs of several genes including MMP9, PTPN1, and SS18L1 were found to be significantly related to metastasis (P<0.05).
Conclusions: We characterized the mutational landscape of 38 GC patients and discovered several potential new predictive markers of survival and metastasis in GC.
Keywords: Whole-exome sequencing (WES); copy number variations (CNV); germline mutation; somatic mutation; tumor mutation burden.
2020 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6620). The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response.JAMA Oncol. 2015 Jul;1(4):466-74. doi: 10.1001/jamaoncol.2015.1313. JAMA Oncol. 2015. PMID: 26181256 Free PMC article.
-
Genomic alterations in gastric cancers discovered via whole-exome sequencing.BMC Cancer. 2018 Dec 19;18(1):1270. doi: 10.1186/s12885-018-5097-8. BMC Cancer. 2018. PMID: 30567531 Free PMC article.
-
Mutational signatures in 175 Chinese gastric cancer patients.BMC Cancer. 2024 Sep 30;24(1):1208. doi: 10.1186/s12885-024-12968-2. BMC Cancer. 2024. PMID: 39350044 Free PMC article.
-
Clinical Interest in Exome-Based Analysis of Somatic Mutational Signatures for Non-Small Cell Lung Cancer.Cancers (Basel). 2024 Sep 9;16(17):3115. doi: 10.3390/cancers16173115. Cancers (Basel). 2024. PMID: 39272973 Free PMC article.
-
An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa.World J Gastroenterol. 2014 Apr 14;20(14):3927-37. doi: 10.3748/wjg.v20.i14.3927. World J Gastroenterol. 2014. PMID: 24744582 Free PMC article. Review.
Cited by
-
Mutation characteristics and molecular evolution of ovarian metastasis from gastric cancer and potential biomarkers for paclitaxel treatment.Nat Commun. 2024 May 4;15(1):3771. doi: 10.1038/s41467-024-48144-0. Nat Commun. 2024. PMID: 38704377 Free PMC article.
-
TIGAR-V2: Efficient TWAS tool with nonparametric Bayesian eQTL weights of 49 tissue types from GTEx V8.HGG Adv. 2021 Nov 4;3(1):100068. doi: 10.1016/j.xhgg.2021.100068. eCollection 2022 Jan 13. HGG Adv. 2021. PMID: 35047855 Free PMC article.
-
Familial Risks and Proportions Describing Population Landscape of Familial Cancer.Cancers (Basel). 2021 Aug 30;13(17):4385. doi: 10.3390/cancers13174385. Cancers (Basel). 2021. PMID: 34503195 Free PMC article.
-
A Deep Neural Network for Gastric Cancer Prognosis Prediction Based on Biological Information Pathways.J Oncol. 2022 Sep 9;2022:2965166. doi: 10.1155/2022/2965166. eCollection 2022. J Oncol. 2022. PMID: 36117847 Free PMC article.
-
Second Primary Cancers After Gastric Cancer, and Gastric Cancer as Second Primary Cancer.Clin Epidemiol. 2021 Jul 2;13:515-525. doi: 10.2147/CLEP.S304332. eCollection 2021. Clin Epidemiol. 2021. PMID: 34239328 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
