HMGB1 as a potential biomarker and therapeutic target for severe COVID-19
- PMID: 33313438
- PMCID: PMC7720697
- DOI: 10.1016/j.heliyon.2020.e05672
HMGB1 as a potential biomarker and therapeutic target for severe COVID-19
Abstract
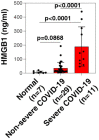
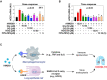
COVID-19 has attracted global attention due to its rapid spread around the world with substantial morbidity and associated mortality. Severe COVID-19 can be complicated by the acute respiratory distress syndrome, sepsis and septic shock leading to death. These complications are thought to result from an overactivation of the immune system, leading to a cytokine storm syndrome associated with multiple organ failure. Here, we report that high mobility group box 1 (HMGB1), a prototypical damage-associated molecular pattern (DAMP) and a central mediator of lethal inflammation, could be a potential target for innovative therapeutic strategies for COVID-19. Serum HMGB1 in severe COVID-19 patients is elevated (189.40 ± 140.88 ng/ml). Exogenous HMGB1 induces the expression of SARS-CoV-2 entry receptor ACE2 in alveolar epithelial cells in an AGER-dependent manner. Importantly, genetic (using AGER siRNA) or pharmacological (using glycyrrhizin, chloroquine, hydroxychloroquine, and FPS-ZM1) inhibition of the HMGB1-AGER pathway blocks ACE2 expression. Thus, HMGB1 inhibitors are likewise promising drug candidates for the treatment of patients suffering from COVID-19.
Keywords: COVID-19; Cell culture; Cell death; HMGB1; Immunology; Infectious disease; Inflammation; Microbiology; Virology.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. - PMC - PubMed
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–575. - PMC - PubMed
-
- Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.D., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H. W.H.O. Strategic, H. Technical advisory group for infectious, COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

