Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection
- PMID: 33314005
- PMCID: PMC7732737
- DOI: 10.1007/s13238-020-00811-w
Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection
Abstract
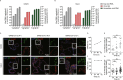
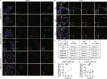
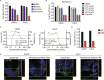
The coronavirus disease 2019 (COVID-19) pandemic is caused by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is spread primary via respiratory droplets and infects the lungs. Currently widely used cell lines and animals are unable to accurately mimic human physiological conditions because of the abnormal status of cell lines (transformed or cancer cells) and species differences between animals and humans. Organoids are stem cell-derived self-organized three-dimensional culture in vitro and model the physiological conditions of natural organs. Here we showed that SARS-CoV-2 infected and extensively replicated in human embryonic stem cells (hESCs)-derived lung organoids, including airway and alveolar organoids which covered the complete infection and spread route for SARS-CoV-2 within lungs. The infected cells were ciliated, club, and alveolar type 2 (AT2) cells, which were sequentially located from the proximal to the distal airway and terminal alveoli, respectively. Additionally, RNA-seq revealed early cell response to virus infection including an unexpected downregulation of the metabolic processes, especially lipid metabolism, in addition to the well-known upregulation of immune response. Further, Remdesivir and a human neutralizing antibody potently inhibited SARS-CoV-2 replication in lung organoids. Therefore, human lung organoids can serve as a pathophysiological model to investigate the underlying mechanism of SARS-CoV-2 infection and to discover and test therapeutic drugs for COVID-19.
Keywords: COVID-19; SARS-CoV-2; cell tropism; cellular metabolism; drug discovery; lung organoids.
© 2020. The Author(s).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

