The Diseased Mitoribosome
- PMID: 33314036
- PMCID: PMC8278227
- DOI: 10.1002/1873-3468.14024
The Diseased Mitoribosome
Abstract
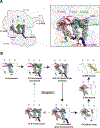
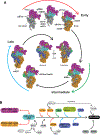
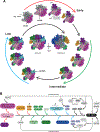
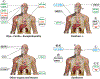
Mitochondria control life and death in eukaryotic cells. Harboring a unique circular genome, a by-product of an ancient endosymbiotic event, mitochondria maintains a specialized and evolutionary divergent protein synthesis machinery, the mitoribosome. Mitoribosome biogenesis depends on elements encoded in both the mitochondrial genome (the RNA components) and the nuclear genome (all ribosomal proteins and assembly factors). Recent cryo-EM structures of mammalian mitoribosomes have illuminated their composition and provided hints regarding their assembly and elusive mitochondrial translation mechanisms. A growing body of literature involves the mitoribosome in inherited primary mitochondrial disorders. Mutations in genes encoding mitoribosomal RNAs, proteins, and assembly factors impede mitoribosome biogenesis, causing protein synthesis defects that lead to respiratory chain failure and mitochondrial disorders such as encephalo- and cardiomyopathy, deafness, neuropathy, and developmental delays. In this article, we review the current fundamental understanding of mitoribosome assembly and function, and the clinical landscape of mitochondrial disorders driven by mutations in mitoribosome components and assembly factors, to portray how basic and clinical studies combined help us better understand both mitochondrial biology and medicine.
Keywords: OXPHOS deficiency; mitochondrial disease; mitochondrial ribosome; mitochondrial translation; mitoribosome assembly.
© 2020 Federation of European Biochemical Societies.
Conflict of interest statement
Conflict of interest statement.
None declared.
Figures

References
-
- McLean JR, Cohn GL, Brandt IK and Simpson MV (1958). Incorporation of labeled amino acids into the protein of muscle and liver mitochondria. J. Biol. Chem. 233, 657–63. - PubMed
-
- Rendi R (1959). On the occurrence of intramitochondrial ribonucleoprotein particles. Exp. Cell Res. 17, 585–7. - PubMed
-
- Kalf GF (1963). The incorporation of leucine-1-C-14 into the protein of rat heart sarcosomes: an investigation of optimal conditions. Arch. Biochem. Biophys. 101, 350–59. - PubMed
-
- Swift H (1965). Nucleic Acids of Mitochondria and Chloroplasts. The American Naturalist 99, 201–214.
-
- Küntzel H and Noll H (1967). Mitochondrial and cytoplasmic polysomes from Neurospora crassa. Nature 215, 1340–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

