Id(entifying) the inhibitor of DNA binding 3 in the brain of Nothobranchius furzeri upon aging
- PMID: 33314133
- PMCID: PMC8053586
- DOI: 10.1111/joa.13367
Id(entifying) the inhibitor of DNA binding 3 in the brain of Nothobranchius furzeri upon aging
Abstract
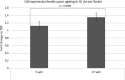
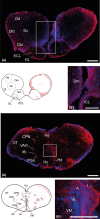
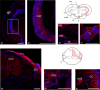
Inhibitors of DNA (Id) are key transcription factors (TFs) regulating neurogenic processes. They belong to the helix-loop-helix (HLH) TF family and are dominant negative regulators of basic HLH proteins (bHLHs). Specifically, they inhibit cell differentiation and enhance cell proliferation and motility. The Id family includes four members, Id1, Id2, Id3, and Id4, which have been identified in nearly all vertebrates. The transcript catalog of the African turquoise killifish, Nothobranchius furzeri, contains all four TFs and has evolved showing positive selection for Id3. N. furzeri, a teleost, is the short-lived vertebrate and is gaining increasing scientific interest as a new model organism in aging research. It is characterized by embryonic diapause, explosive sexual maturation, and rapid aging. In this study, we investigated both the expression and the role of Id3 in the brain of this model organism. Interestingly, Id3 was upregulated age-dependently along with a distribution pattern resembling that of other vertebrates. Additionally, the gene has undergone positive selection during evolution and shows a high degree of conservation relative to that of other vertebrates. These features make N. furzeri a valid tool for aging studies and a potential model in translational research.
Keywords: Id3; aging; central nervous system; fish.
© 2020 Anatomical Society.
Conflict of interest statement
None declared.
Figures


References
-
- Afouda, A.B. , Reyonaud‐Deonauth, S. , Mohun, T. & Spohr, G. (1999) Localized XId3 mRNA activation in Xenopus embryos by cytoplasmatic polyadenylation. Mechanisms of Development, 88(1), 15–31. - PubMed
-
- Andres‐Barquin, P. , Hernandez, M.C. & Israel, M.A. (2000) Id genes in nervous system development. Histology and Histopathology, 15(2), 603–618. - PubMed
-
- Benezra, R. (1993) Id as a general negative regulator of the helix‐loop‐helix family of DNA‐binding proteins. Pharmacology of Cell Differentiation, 1016, 171–180.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

