Computational drug repurposing study of the RNA binding domain of SARS-CoV-2 nucleocapsid protein with antiviral agents
- PMID: 33314794
- PMCID: PMC7883068
- DOI: 10.1002/btpr.3110
Computational drug repurposing study of the RNA binding domain of SARS-CoV-2 nucleocapsid protein with antiviral agents
Abstract
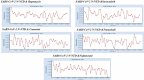
The recent outbreak of coronavirus disease (COVID-19) in China caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to worldwide human infections and deaths. The nucleocapsid (N) protein of coronaviruses (CoVs) is a multifunctional RNA binding protein necessary for viral RNA replication and transcription. Therefore, it is a potential antiviral drug target, serving multiple critical functions during the viral life cycle. This study addresses the potential to repurpose antiviral compounds approved or in development for treating human CoV induced infections against SARS-CoV-2 N. For this purpose, we used the docking methodology to better understand the inhibitory mechanism of this protein with the existing 34 antiviral compounds. The results of this analysis indicate that rapamycin, saracatinib, camostat, trametinib, and nafamostat were the top hit compounds with binding energy (-11.87, -10.40, -9.85, -9.45, -9.35 kcal/mol, respectively). This analysis also showed that the most common residues that interact with the compounds are Phe66, Arg68, Gly69, Tyr123, Ile131, Trp132, Val133, and Ala134. Subsequently, protein-ligand complex stability was examined with molecular dynamics simulations for these five compounds, which showed the best binding affinity. According to the results of this study, the interaction between these compounds and crucial residues of the target protein were maintained. These results suggest that these residues are potential drug targeting sites for the SARS-CoV-2 N protein. This study information will contribute to the development of novel compounds for further in vitro and in vivo studies of SARS-CoV-2, as well as possible new drug repurposing strategies to treat COVID-19 disease.
Keywords: COVID-19; CoVs; MD simulations; N protein; RNA binding domain; SARS-CoV-2; drug repurposing; molecular docking.
© 2020 American Institute of Chemical Engineers.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures



References
-
- Gorbalenya AE, Snijder EJ, Ziebuhr J. Virus‐encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81(4):853‐879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

