Influence of vintage, geographic location and cultivar on the structure of microbial communities associated with the grapevine rhizosphere in vineyards of San Juan Province, Argentina
- PMID: 33315910
- PMCID: PMC7735631
- DOI: 10.1371/journal.pone.0243848
Influence of vintage, geographic location and cultivar on the structure of microbial communities associated with the grapevine rhizosphere in vineyards of San Juan Province, Argentina
Abstract
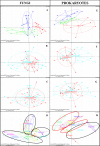
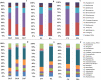
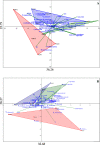
Soil microbiomes, as a primary reservoir for plant colonizing fungi and bacteria, play a major role in determining plant productivity and preventing invasion by pathogenic microorganisms. The use of 16S rRNA and ITS high-throughput amplicon sequencing for analysis of complex microbial communities have increased dramatically in recent years, establishing links between wine specificity and, environmental and viticultural factors, which are framed into the elusive terroir concept. Given the diverse and complex role these factors play on microbial soil structuring of agricultural crops, the main aim of this study is to evaluate how external factors, such as vintage, vineyard location, cultivar and soil characteristics, may affect the diversity of the microbial communities present. Additionally, we aim to compare the influence these factors have on the structuring of bacterial and fungal populations associated with Malbec grapevine rhizosphere with that of the more widespread Cabernet Sauvignon grapevine cultivar. Samples were taken from Malbec and Cabernet Sauvignon cultivars from two different vineyards in the San Juan Province of Argentina. Total DNA extracts from the rhizosphere soil samples were sequenced using Illumina's Miseq technology, targeting the V3-V4 hypervariable 16S rRNA region in prokaryotes and the ITS1 region in yeasts. The major bacterial taxa identified were Proteobacteria, Bacteroidetes and Firmicutes, while the major fungal taxa were Ascomycetes, Basidiomycetes, Mortierellomycetes and a low percentage of Glomeromycetes. Significant differences in microbial community composition were found between vintages and vineyard locations, whose soils showed variances in pH, organic matter, and content of carbon, nitrogen, and absorbable phosphorus.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Del Frari G, Gobbi A, Aggerbeck MR, Oliveira H, Hansen LH, Ferreira RB (2019a) Characterization of the wood mycobiome of Vitis vinifera in a vineyard affected by Esca. spatial distribution of fungal communities and their putative relation with leaf symptoms. Frontiers in plant science 10: 910 10.3389/fpls.2019.01405 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

