Deep Learning Predicts Underlying Features on Pathology Images with Therapeutic Relevance for Breast and Gastric Cancer
- PMID: 33316873
- PMCID: PMC7763049
- DOI: 10.3390/cancers12123687
Deep Learning Predicts Underlying Features on Pathology Images with Therapeutic Relevance for Breast and Gastric Cancer
Abstract
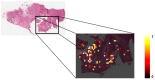
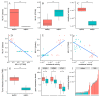
DNA repair deficiency (DRD) is an important driver of carcinogenesis and an efficient target for anti-tumor therapies to improve patient survival. Thus, detection of DRD in tumors is paramount. Currently, determination of DRD in tumors is dependent on wet-lab assays. Here we describe an efficient machine learning algorithm which can predict DRD from histopathological images. The utility of this algorithm is demonstrated with data obtained from 1445 cancer patients. Our method performs rather well when trained on breast cancer specimens with homologous recombination deficiency (HRD), AUC (area under curve) = 0.80. Results for an independent breast cancer cohort achieved an AUC = 0.70. The utility of our method was further shown by considering the detection of mismatch repair deficiency (MMRD) in gastric cancer, yielding an AUC = 0.81. Our results demonstrate the capacity of our learning-base system as a low-cost tool for DRD detection.
Keywords: DNA repair deficiency; biomarker; deep learning; digital pathology; mutational signature.
Conflict of interest statement
The authors declare no conflict of interest
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

